ABSTRACT
Escherichia coli and Klebsiella pneumoniae are pathogens frequently involved in urinary tract infections with high epidemic potential. The increase and spread resistance of these microbes to broad spectrum beta-lactam antibiotics are usually reported and is a real public health concern in Côte d’Ivoire but information on genetic variants and intragenic mutations encoding these resistances are scarce. The aim of this study is to characterize genetic variants and describe the intragenic mutations underlying resistance to broad-spectrum beta-lactam antibiotics in uropathogen E. coli and K. pneumoniae in HKB and CHR hospitals with different epidemiological facies in Côte d’Ivoire. 39 strains comprising 30 of E. coli and 9 strains of K. pneumoniae were isolated from which DNA was extracted, amplified and sequenced. ESBLs genes were detected by polymerase chain reaction in 58.8 % of strain analysis. No significant difference was observed between ESBL from HKB and CHR hospitals although HKB and CHR sites present 50 and 56.8% of ESBL respectively. Nucleotide sequences subjected to BLASTn for sequences similarity and homology revealed diversity of resistance genes with dominance of the gene encoding the extended-spectrum β-lactamase CTX-M-15 and the emergence of a new blaTEM-9 gene in Côte d'Ivoire. The significant co-expression of ESBLs might impact 3rd generation cephalosporin multi-resistance among pathogenic bacteria infecting patient population. Routine antibiogram practice could guide the choice of optimal antibiotic therapy for successful treatment and delay the occurrence of multidrug resistance in enterobacterial infections.
Key words: Urinary tract infection, extended-spectrum β-lactamase, gene variants, mutations, antibiotic resistance, Côte d’Ivoire.
Escherichia coli and Klebsiella pneumoniae are commensal enterobacteria of the intestinal microflora of humans and warm-blooded animals (Ardakani and Ranjbar, 2016). These microbes are ability used to colonize the extra-intestinal tracts of the host by acquisition virulence factors and caused a pathological state in this one (Massot et al., 2016). Several studies showed that E. coli and K. pneumoniae are the pathogens frequently involved in hospital and community-acquired urinary tract infections (Hyun et al., 2019; Yadav et al., 2019). Because of their high frequency in infections, they are constantly subjected to antibiotics used in irrational, empirical and probabilistic ways (Jena et al., 2017) and broad-spectrum antibiotics, mainly β-lactam molecules, are the most recommended by clinicians (Lima et al., 2020). Given its low cost and availability which result in over use (Ventola, 2015), β-lactam antibiotic experienced resistance leading to therapeutic failures (Friedman et al., 2016). Indeed, bacteria, in a hostile environment generated by drug pressure, develop resistance mechanisms or acquire mobile genetic elements (Munita and Arias, 2016). The most common mechanism of resistance in Enterobacteriaceae is related to expression of extended-spectrum beta lactamases (ESBLs) (Shaikh et al., 2015; Teklu et al., 2019). This resistance now extends to the latest generation of cephalosporins and carbapenems (Kim et al., 2017). blaTEM, blaSHV and blaCTX-M are the main variants of genes coding for resistance in Enterobacteriaceae with the latest one (Ahmad and Khadija, 2019). In Côte d'Ivoire, recent studies have reported the spread of ESBL-producing strains of E. coli and K. pneumoniae in various infections (Tahou et al., 2017; Gadou et al., 2018) although the setup since 2002 of an observatory of antimicrobial resistance is dedicated to actions contributing to fight against the spread of resistant or multi-resistant bacteria (Ouédraogo et al., 2017). In addition, information on genetic variants and intragenic mutations encoding these resistances are scarce. This study aim to determine gene variants as well as nucleotide and peptide mutations involved in resistance to broad-spectrum beta-lactam antibiotics in uropathogenic E. coli and K. pneumoniae in Côte d’Ivoire.
Study sites description
This study was carried out in two hospitals belonging to different epidemiological facies: the Henriette Konan Bedié Hospital in Abobo (HKB-Abobo) located in Abidjan 03'86'501 N; 06'01'275 and the Regional Hospital Center of Haut-Sassandra (CHR-Daloa) located in Daloa 07'82'734 N; 07'61'563 W (Figure 1). At HKB hospital, medical management of bacterial infections is carried out according to an empirical and probabilistic model due to the lack of antibiograms implementation which could lead to an accurate diagnosis and the prescription of the correct antibiotic. In addition, this hospital is located in one of the most populated municipalities of Abidjan, with an estimated population of about 1.03 million inhabitants (RGPH, 2014) with common practice of self-medication. These factors actively contribute to the spread of a wide variety of bacterial pathogens that are resistant to a large number of antibiotics. CHR hospital in Daloa remains the only hospital in the Haut-Sassandra region which has a medical microbiology laboratory which carries out routinely microbial tests. Consequently, it receives patients from all four geographical points of the region for bacterial infection cases, showing a good distribution of bacterial infection cases in the region.
Ethics statement
The study protocol was reviewed and approved by the national ethics committee of Life Sciences and Health in Côte d’Ivoire with the number: N/Ref:106-18/MSHP/ CNESVS-KM, US DPT OF HHS REGISTRATION $: IORG00075 on 30th July 2018. This study is part of the ESTHER project which aims to do a better diagnosis of urinary tract infections in outpatients in addition to microbial surveillance of uropathogens and antimicrobial resistance. Consent was obtained from patients and/or guardians after explaining the objective of the study. The laboratory results were communicated to patients via physicians for better antibiotic prescription.
Study design and sample collection
The sampling of this prospective and descriptive study was performed from October 2018 to April 2019 at HKB and from May to October 2019 at CHR Daloa. During this period, a total of 39 strains including 30 strains of E. coli and 9 strains of K. pneumoniae were isolated from fresh urine samples from patients with urinary tract infections.
Isolation and identification of bacterial strains
The uropathogenic bacterial strains were isolated on CHROMAgar Orientation (Becton Dickinson, Cockeysville, MD), chromogenic medium (Manickam et al., 2013). The identification of E. coli and K. pneumoniae species was performed using Gram staining tests and classical biochemical tests such as indole, oxidase, catalase, urease, tryptophan deaminase, glucose and lactose fermentation, production of gases from glucose fermentation, degradation of hydrogen peroxide by the production of hydrogen sulfide, use of citrate as the unique source of carbon, motility, lysine deaminase and lysine decarboxylase production (Tandon and Bhargava, 2019). Samples collection details are presented by the algorithm in Figure 2.
Phenotypic detection of uropathogenic ESBL-producing strains
The uropathogenic E. coli and K. pneumoniae strains, producing extended-spectrum beta-lactamases (ESBL) were phenotypically detected by double synergistic method on Müller Hinton agar (Jarlier et al., 2000). This testing consisted in appearance of "champagne cork" characteristic image within an antibiotic disc containing a beta-lactamase inhibitor such as amoxicillin + clavulanic acid (AMC) and third-generation cephalosporin discs such as ceftazidime (CAZ), ceftriaxone (CRO), aztreonam (ATM) and cefotaxime (CTX).
Molecular characterization of ESBL-producing strains
DNA extraction
The extraction of genomic DNA from E. coli and K. pneumoniae strains was performed using the phenol-chloroform method described by Chan and Goodwin (1995).
Genotypic detection of ESBLs
The following primers blaTEM, blaSHV and blaCTX-M were used to amplify specific DNA sequences involved in resistance to broad-spectrum beta-lactam antibiotics by PCR (Table 1). Total reaction volume of 50 μl containing 1 μl of each primer of 10 pmol/μl (Eurogentec, Blegium), 5 μl of MgCl2 PCR buffer 10 x (Qiagen), 2.5 μl of deoxyribonucleoside triphosphates (dNTPs, 200 μM), 0.1 μl of Taq polymerase (Qiagen), 37.4 μl of ultrapure water and 3 μl of bacterial genomic DNA was mixed. This PCR mixture was performed in a thermal cycler under the following conditions: initial denaturation for 5 min at 94°C followed by 30 cycles consisting of denaturation at 94°C for 45 s, hybridization at 60°C for 1 min and elongation at 72°C for 1 min followed by final elongation at 72°C for 10 min.
Sequencing of ESBL resistance genes
Bidirectional sequencing of BLSE-positive PCR products (blaTEM, blaCTX-M and blaSHV) was performed by BGI TECH SOLUTIONS (HONG-KONG) using the ABI PRISM 3730 automated sequencer (Applied Biosystems).
Bioinformatics analysis
Nucleotide sequences were compared to reference sequences in Genbank genomic database of National Center for Biotechnology Information (NCBI) using the BLASTN local alignment search tool available online (http://www.ncbi.nih.gov). Protein sequences derived from the genes were aligned using DNA Baser Assembler 5.15.0 and analyzed in order to identify the CTX-M and TEM type according to sequences leading to detection of the mutations underlying resistance to ESBLs. The statistical Chi-square test of independence and the Fisher exact test were performed to compare the proportions of ESBLs strains. Significant differences are observed when the probability value (p) associated with the statistical tests is strictly less than 0.05.
Phenotypic determination of ESBLs
Figure 3 shows the phenotypic detection of a broad- spectrum beta-lactamase-producing bacterial strain using the double-synergy method on Müller Hinton agar. ESBLs production is detected by the appearance of a particular champagne cork image. The distribution of broad-spectrum beta-lactamase-producing isolates is indicated in Table 2. Results showed that 21 (53.8%) out of 39 study strains produced ESBL. E. coli and K. pneumoniae species expressed 56.7 and 44.4% at least one ESBL respectively. At the HKB hospital in Abobo, 50 % of strains analyzed were BLSE-producing positive, while this phenotype was expressed in 58.8 % of the strains at the CHR-Daloa. Although the prevalence of ESBL strains is slightly higher at CHR hospital in Daloa, the difference was not statistically significant (P = 0.82). This result showed that ESBL production was not significantly associated with bacterial specie in this study (Table 2).
Molecular detection of ESBLs resistance
The genes coding for resistance to ESBL, namely blaTEM, blaSHV and blaCTX-M were tested for the 21 strains with the ESBL phenotype. The electrophoretic profiles of the detected BLSE genes are shown in Figure 4. Genotyping revealed the presence of two types of ESBL genes: The blaTEM gene encoding the TEM enzyme and the blaCTX-M gene encoding the CTX-M enzyme. The blaTEM and blaCTX-M genes are characterized by DNA fragments of 112 and 701 base pairs respectively (Figure 4a and b), but the blaSHV gene was not detected in this study.
Distribution of broad-spectrum beta-lactamases resistance genes in E. coli and K. pneumonia
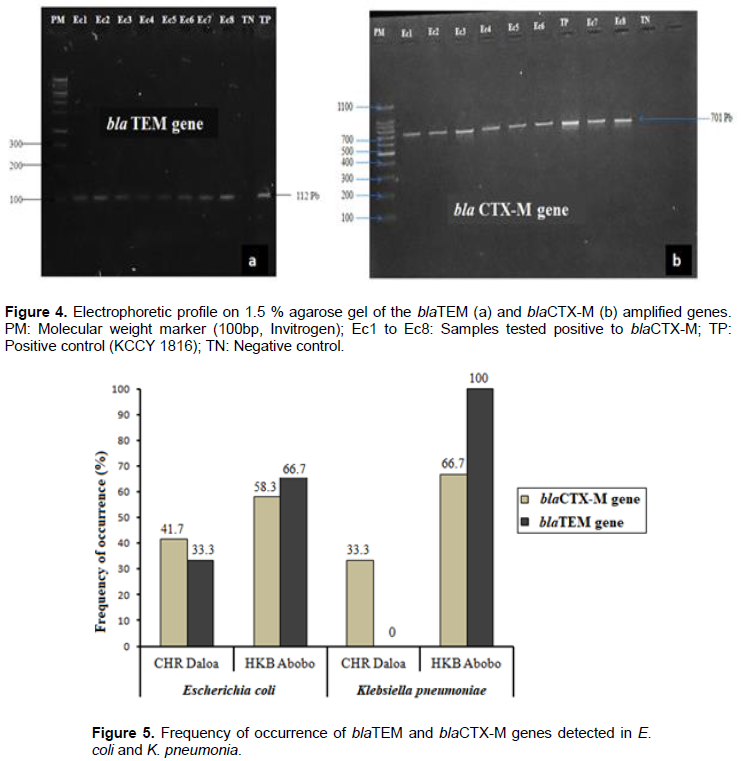
The distribution of the blaCTX-M and blaTEM genes encoding resistance to the broad-spectrum beta-lactamases detected in E. coli and K. pneumoniae in this study are shown in Figure 5. In the population of E. coli producing extended-spectrum beta-lactamase, the blaTEM gene was detected in nine strains (52.94 %), including six strains from HKB Abobo Hospital (66.7%) and three strains (33.3%) from CHR Daloa. Concerning the blaCTX-M gene, an overall frequency of occurrence of 70.6% is obtained in these bacterial species. In the collection from Abobo HKB hospital, the proportion of E. coli strains expressing the blaCTX-M gene was 58.3% (7/12), while at Daloa CHR, the proportion was 41.7%(5/12). Among the four ESBL-producing strains of K. pneumoniae, an overall occurrence frequency of 75 % (3 strains) is obtained for the blaTEM gene. This gene was detected in all ESBL-producing strains from HKB hospital in Abobo (100%). However, K. pneumoniae strain isolated at CHR Daloa did not carry the blaTEM gene. The blaCTX-M gene was detected in 3 strains of which two were from HKB hospital in Abobo. The unique strain of K. pneumoniae collected at CHR Daloa was a carrier of the blaCTX-M gene, with 33.3% frequency of occurrence. In addition, co-occurrences for these two genes (blaTEM and blaCTX-M) were observed in this study in four strains of E. coli (66.7%) and two strains of K. pneumoniae (33.3%) (Figure 5).
Bioinformatics Analysis of blaTEM and blaCTX-M genes
Out of twenty-one strains expressing ESBL genes detected and sent for sequencing, fifteen were successfully sequenced. These strains consisted of three strains of K. pneumoniae and twelve strains of E. coli. Analysis of TEM and CTX-M genes sequences were done to determine their relationship to other TEM and CTX-M gene sequences available in Gen-Bank database using BLAST nucleotide algorithm (http://www.ncbi.nlm.nih.gov/). Variants of the blaTEM sequences analysis revealed that among the seven strains (E. coli1, E. coli5, E. coli14, E. coli6, E. coli11, E. coli9 and Kleb2), only one strain (14.3 %) isolated at HKB hospital produced the TEM-1 enzyme (Table 3). The corresponding nucleotide sequence was identified to be 97 % homologous to the coding sequence of the 66 base pair length TEM-1 allele, available in the Genbank under accession number KT415643. Alignment of this sequence revealed two base point mutations with changes in amino acid at position 38; the adenine of the CCA codon in the reference sequence was substituted by cytosine (A38C) and at position 48 the guanine of the TGT codon in the reference sequence was substituted by thymine (G48T). These nucleotide mutations induced an amino acid change in the translated peptide sequence. Accordingly, at position 13 of this sequence, glutamine mutated to proline (Gln13Pro) and at position 16 tryptophan mutated to cysteine (Trp16Cys) in the newly formed sequence (Figure 6).
Apart from TEM-1 enzyme, the others strain 85.7% revealed the production of the TEM-9 enzyme with 95.2 to 100 % similarity to the reference TEM-9 allele (accession number KY271103) available in Genbank. As illustration, the nucleotide sequence of the E. coli _6 isolate shows 98.6 % homology with the reference TEM-9 allele of the blaTEM gene (Figure 7, Table 3). A point mutation of bases was detected in position 3 regarding nucleotide alignment where A base in reference sequence was replaced by a C in the query sequence. However, after translation into amino acid sequences, no mutation was observed (Figure 7).
With the E. coli_1 strain, alignment results showed a 95.2% of similarity between the nucleotide sequence of the blaTEM gene and the reference sequence coding for the TEM-9 allele (accession number KY271103). The alignment of these two sequences revealed a substitution at position 4 of the thymine TGT codon by adenine (T4A) in the query sequence. A gap extension at positions10 and 11 and an insertion of cytosine at position 55 were also observed in E. coli_1 strain sequence (Figure 7). The peptide sequence resulting from this alignment showed numerous amino acid changes from position 2 to 19 and from 21 to 27 (Figure 8). For E. coli_5, E. coli_11, E. coli_14 and Kleb_2 isolates a 100% similarity was obtained with the reference TEM-9 allele of the blaTEM gene accessible at KY271103 in the Genbank. The TEM-1 and TEM-9 variants of the blaTEM gene detected in this study are mainly produced by strains from HKB hospital.
Alignment of the nucleotide sequences of the blaCTX-M gene of the studied strains revealed a single variant of this gene, CTX-M-15 with 100 % identity to the coding reference sequence (accession number MN816278) in Genbank (Figure 9). CTX-M-15 is the only CTX-M-like enzyme produced by the studied strains to resist broad-spectrum beta-lactams. This variant was detected in 10 uropathogenic strains (66.7%) versus seven strains (46.7%) for the blaTEM variant. These strains comprised one strain of K. pneumoniae from each of the two study sites and eight strains of E. coli (four from HKB hospital in Abobo and four from the CHR hospital in Daloa).
Faced with drug pressure, bacteria develop resistance mechanisms whose expression is encoded by genetic factors subject to mutations. In this study, 53.8% of strains analyzed produced at least one broad-spectrum beta-lactamase. Of the two bacteria species studied. E. coli is the one which presents the greatest prevalence of resistance (56.7%) even if the 44% presented by K. pneumoniae are not negligible. The high prevalence of ESBLs in this study, as well as some reported in Côte d’ d’Ivoire (Guessennd et al., 2011; Gadou et al.. 2018) and in other parts of Africa (Togo et al., 2014; Saravanan et al., 2018; Ouchar et al., 2019), show that the spread of ESBL-producing Enterobacteriaceae in an African context remains a very worrisome phenomenon. The high proportion of uropathogenic strains producing ESBL is thought to be related to the widespread use of third-generation cephalosporins and lack of adequate urinary tract infection (UTI) treatment regarding dosing regimens (Halaji et al., 2020). Indeed, the prevalence of the resistant phenotype is often an accurate reflection of antibiotic prescribing habits (Llor and Bjerrum, 2014). The molecular profile of the 21 strains is characterized by the expression of two main types of ESBL enzymes, namely CTX-M and TEM. The blaCTX-M gene was the most involved in ESBL resistance in E. coli, while, blaTEM gene was detected at 75% in ESBL resistance in K. pneumoniae. Expression of these genes may be the cause of failures observed in UTI treatment, particularly using extended-spectrum beta-lactam drugs (Hagel et al., 2013). The concomitant expression of blaTEM and blaCTX-M genes revealed in this study could be due to cross-resistance to penicillins and acquired cephalosporins, suggesting the multi-resistant character of these strains (Ahmad and Khadija, 2019).
In silico analysis of the DNA and protein sequences of the blaTEM gene revealed two enzyme variants, TEM-1 and TEM-9. The TEM-1 variant observed with proline/glutamine substitutions is an original plasmid-mediated beta-lactamase from which all other TEM variants are derived by mutation of one or more amino acids (Ur Rahman et al., 2018). Commonly found in E. coli, TEM-1 is able to hydrolyze penicillins and narrow spectrum cephalosporins such as cephalothin (Ur Rahman et al., 2018; Galindo- Méndez, 2020). TEM-1 has also been reported in Côte d'Ivoire from previous studies (Guessennd et al. 2008; Tahou et al., 2017) in Klebsiella pneumoniae. According to the scientific literature, the TEM-9 variant is detected for the first time in Côte d'Ivoire. This variant was derived from TEM-1 by four amino acid substitutions and was first characterized by Mabilat et al. (1990) in a clinical strain of K. pneumoniae at the Royal Hospital in Hallamshire in England. This mutation leads to resistance to third-generation cephalosporins.particularly ceftazidime (Jahani et al., 2017). In addition to blaTEM, bla CTX-M was also detected with the CTX-M-15 variant being the only one observed in this study. Initially reported in India in 1999, this variant predominates the current molecular epidemiology of ESBLs in K. pneumoniae and E. coli involved in both community UTI and nosocomial infections (Chong et al., 2018). The co-expression of the TEM-9 and CTX-M-15 variants, observed in the E. coli1_1 strain in this study is evidence of the multi-drug resistant nature of the strains circulating in the African region, due to the ability of this organism to hydrolyze larger substrates and an extended transmission of these genetic resistance factors (Parajuli et al., 2016).
This study showed a high proportion of ESBL-producing strains in UTI involving E. coli and K. pneumoniae species. Molecular epidemiology and in silico analysis of antibiotic resistance in these two uropathogens is characterized by a diversity of beta-lactam resistance genes with the emergence for the first time of blaTEM-9 gene variant in Côte d'Ivoire and the dominance of the gene encoding ESBL CTX-M-15, confirming the high diffusion of this variant worldwide. Furthermore, the study highlighted significant co-expression of ESBLs imparting 3rd generation cephalosporin resistance among pathogenic bacteria infecting patient population. Routine antibiogram practice could guide the choice of optimal antibiotic therapy for successful treatment and delay the occurrence of multidrug resistance in Enterobacterial infections.
The authors have not declared any conflict of interests.
The authors appreciate M. Yeo from HKB Hospital and Dr Monto from CHR Hospital for their support in sample collection. Our thanks also go to the ESTHER project for financial support.
REFERENCES
|
Ahmad HP, Khadija KM (2019). Prevalence of blaTEM, blaSHV, and blaCTX-M Genes among ESBL-Producing Klebsiella pneumoniae and Escherichia coli Isolated from Thalassemia Patients in Erbil, Iraq. Mediterranean Journal of Hematology and Infectious Diseases 11(1):1-7.
Crossref
|
|
|
|
Ardakani MA, Ranjbar R (2016). Molecular typing of uropathogenic E. coli strains by the ERIC-PCR method. Electronic Physician 8(4):2291-2296.
Crossref
|
|
|
|
|
Chan JW, Goodwin PH (1995). Extraction of genomic DNA from extracellular polysaccharide-synthesizing gram negative bacteria. Biotechniques 18(3):418-422.
|
|
|
|
|
Chong Y, Shimoda S, Shimono (2018). Current epidemiology genetic evolution and clinical impact of extended-spectrum b-lactamase-producing Escherichia coli and Klebsiella pneumoniae. Infection Genetics and Evolution 61:185-188.
Crossref
|
|
|
|
|
Friedman ND, Temkin E, Carmeli Y (2016). The negative impact of antibiotic resistance. Clinical Microbiology and Infection 22(5):416-422.
Crossref
|
|
|
|
|
Gadou V, Guessennd NK, Toty AA, Konan F, Ouattara MB, Dosso M, Diène SM, Djaman JA, Rolain JM (2018). Molecular Detection of the Arr-2 Gene in Escherichia coli and Klebsiella pneumoniae Resistant to Rifampicin in Abidjan. Côte D'Ivoire. Microbiology Research Journal International 23(4):1-8.
Crossref
|
|
|
|
|
Galindo-Méndez M (2020). Antimicrobial Resistance in Escherichia coli. E. Coli Infections - Importance of Early Diagnosis and Efficient Treatment pp. 1-20.
Crossref
|
|
|
|
|
Guessennd N, Bremont S, Gbonon V, Kacou-NDouba A, Ekaza E, Lambert T, Dosso M, Courvalin P (2008). Résistance aux quinolones de type qnr chez les entérobactéries productrices de bêta-lactamases à spectre élargi à Abidjan en Côte d'Ivoire. Pathologie Biologie 56:439-446.
Crossref
|
|
|
|
|
Guessennd N, Gbonon V, Ouattara ND, Kacou NA, Ekaza E, Yapi D, Affi MR, Guinan JC, Dosso M, Courvalin P (2011). Entérobactéries productrices de B-lactamases à spectre élargi (BLSE) d'origine communautaire à Abidjan de 2005 à 2006: prévalence et niveau de résistance aux antibiotiques. Revue BioAfrica 9:5-11.
|
|
|
|
|
Hagel S, Stallmach A, Keller P, Pletz M (2013). Multiresistant Organisms. Zentralblatt Fur Chirurgie 140(4):417-425.
Crossref
|
|
|
|
|
Halaji M, Shahidi S, Atapour A, Ataei B, Feizi A, Havaei SA (2020). Characterization of Extended-Spectrum β-Lactamase-Producing Uropathogenic Escherichia coli Among Iranian Kidney Transplant Patients. Infection and Drug Resistance13:1429-1437.
Crossref
|
|
|
|
|
Hyun M, Lee JY, Kim HA, Ryu SY (2019). Comparison of Escherichia coli and Klebsiella pneumonia Acute Pyelonephritis in Korean Patients. Infection & Chemotherapy 51(2):130-141.
Crossref
|
|
|
|
|
Jahani S, Ghamgosha M, Shakiba A, Hassanpour K, Taheri RA, Farnoosh G (2017). Assessment of Third Generation Cephalosporin (Ceftazidime and Ceftriaxone) Resistant Escherichia Coli Strains Isolated from Zahedan Hospitals by Tracing the TEM Gene. Journal of Applied Biotechnology Reports 4(1):547-552.
|
|
|
|
|
Jarlier V, Nordmann P, Freney J, Renaud F, Hansen W, Botler C (2000). Entérobactéries et bêta-lactamines. In:Précis de bactériologie clinique. ESKA, Paris (France) pp. 649-665.
|
|
|
|
|
Jena J, Debata NK, Saho RK, Gaur M, Subudhi E (2017). Genetic diversity study of various β-lactamase-producing multidrug-resistant Escherichia coli isolates from a tertiary care hospital using ERIC-PCR. Indian Journal of Medical Research 146(7):23-29.
Crossref
|
|
|
|
|
Kim D, Ahn JY, Lee CH, Jang SJ, Lee H, Yong D, Jeong SH, Lee K (2017). Increasing Resistance to Extended-Spectrum Cephalosporins, Fluoroquinolone and Carbapenem in Gram-Negative Bacilli and the Emergence of Carbapenem Non-Susceptibility in Klebsiella pneumoniae: Analysis of Korean Antimicrobial Resistance Monitoring System (KARMS) Data from 2013 to 2015. Annals of Laboratory Medicine 37(3):231-239.
Crossref
|
|
|
|
|
Lima LM, Monteiro da Silva BN, Barbosa G, Barreiro EJ (2020). β-lactam antibiotics: An overview from a medicinal chemistry perspective. European Journal of Medicinal Chemistry pp. 1-72.
Crossref
|
|
|
|
|
Llor C, Bjerrum L (2014). Antimicrobial resistance: risk associated with antibiotic overuse and initiatives to reduce the problem. Therapeutic Advances in Drug Safety 5(6):229-241.
Crossref
|
|
|
|
|
Mabilat C, Goussard S, Sougakoff W, Spencer R C, Courvalin P (1990). Direct sequencing of the amplified structural gene and promoter for the extended-broad-spectrum β-lactamase TEM-9 (RHH-1) of Klebsiella pneumoniae. Plasmid 23(1):27-34.
Crossref
|
|
|
|
|
Manickam K, Karlowsky JA, Adam H, Lagace-Wiens PRS, Rendina A, Pang P, Murray BL, Alfa MJ (2013). CHROMagar Orientation Medium Reduces Urine Culture Workload. Journal of Clinical Microbiology 51(4):1179-1183.
Crossref
|
|
|
|
|
Massot M, Picard B, Denamur E (2016). Diversité des populations d'Escherichia coli et leurs variations au cours du temps au sein du microbiote intestinal. Revue Francophone Des Laboratoires 486:35-43.
Crossref
|
|
|
|
|
Munita JM, Arias CA (2016). Mechanisms of antibiotic resistance. Microbiol Spectrum 4(2):481-511.
Crossref
|
|
|
|
|
Ouchar MO, Lounnas M, Hide M, Dumont Y, Tidjani A, Kamougam K, Abderrahmane M, Benavides J, Solassol J, Bañuls AL, Jean-pierre H, Carriere C, Godreuil S (2019). High prevalence and characterization of extended-spectrum ß-lactamase producing Enterobacteriaceae in Chadian hospitals. BMC Infectious Diseases 19(1):1-7.
Crossref
|
|
|
|
|
Ouédraogo AS, Jean-Pierre H, Banuls AL, Ouédraogo R, Godreuil S (2017). Emergence et diffusion de la résistance aux antibiotiques en Afrique de l'Ouest : facteurs favorisants et évaluation de la menace. Médecine et Santé Tropicales 27:147-154.
Crossref
|
|
|
|
|
Parajuli NP, Maharjan P, Joshi G, Khanal P R (2016). Emerging Perils of Extended Spectrum β-Lactamase Producing Enterobacteriaceae Clinical Isolates in a Teaching Hospital of Nepal. BioMed Research International pp. 1-7.
Crossref
|
|
|
|
|
RGPH (2014). Resultats globaux:
View. 22 p [Accessed June 07. 2020].
|
|
|
|
|
Saravanan M, Ramachandran B, Barabadi H (2018). The prevalence and drug resistance pattern of extended spectrum β-lactamases (ESBLs) producing Enterobacteriaceae in Africa. Microbial Pathogenesis 114:180-192.
Crossref
|
|
|
|
|
Shaikh S, Fatima J, Shakil S, Rizvi SMD, Kamal M (2015). Antibiotic resistance and extended spectrum beta-lactamases: Types. epidemiology and treatment. Saudi Journal of Biological Sciences 22(1):90-101.
Crossref
|
|
|
|
|
Tahou EJ, Guessesennd KN, Gba KMK, Gbonon V, Konan F, Tiekoura BK, Ouattara MB, Toti A, Kouadio NJ, N'Guetta AS (2017). Characterization and horizontal transfert of enhanced spectrum beta-lactamases production in Klebsiella pneumoniae clinical strains from 2011 to 2016 in Abidjan (Côte d'Ivoire). International Journal of Current Advanced Research 6(12):8118-8122.
|
|
|
|
|
Tandon N, Bhargava B (2019). Standard Operating Procedure Bacteriology. Antimicrobial Resistance surveillance and Research Network. Indian Council of Medical Research (New Delhi), India 199 p.
|
|
|
|
|
Teklu DS, Negeri AA, Legese MH, Bedada TL, Woldemariam HK, Tullu KD (2019). Extended-spectrum beta-lactamase production and multi-drug resistance among Enterobacteriaceae isolated in Addis Ababa. Ethiopia Antimicrobial Resistance and Infection Control 8(39):1-12.
Crossref
|
|
|
|
|
Togo Y, Kubo T, Taoka R, Hiyama Y, Uehara T, Hashimoto J, Kurimura Y, Takahashi S, Tsukamoto T, Miyazaki J, Nishiyama H, Kira S, Kiyota H, Yazawa S, Niwa N, Hongo H, Oya M, Kato T, Yasuda M, Deguchi T, Ishikawa K, Hoshinaga K, Matsumoto M, Shigemura K, Tanaka K, Arakawa S, Fujisawa M, Wada K, Uehara S, Watanabe T, Kumon H, Kobayashi K, Matsubara A, Matsumoto M, Sho T, Hamasuna R, Matsumoto T, Hayami H, Nakagawa M, Yamamoto S (2014). Occurrence of infection following prostate biopsy procedures in Japan: Japanese Research Group for Urinary Tract Infection (JRGU) - a multi-center retrospective study. Journal of Infection and Chemotherapy 20(4):232-237.
Crossref
|
|
|
|
|
Ur Rahman S, Ali T, Ali I, Khan NA, Han B, Gao J (2018). The Growing Genetic and Functional Diversity of Extended Spectrum Beta-Lactamases. BioMed Research International pp. 1-14.
Crossref
|
|
|
|
|
Ventola CL (2015). The antibiotic resistance crisis: part 1: causes and threats. peer-reviewed journal for formulary management 40(4):277-283.
|
|
|
|
|
Yadav MK, Maheshwari RK, Mishra RK, Malhotra B (2019). Isolation and antibiotic susceptibility of Escherichia coli from urine sample received in SMS Medical College, Jaipur. International Journal of Medical and Health Research 5(4):5-9.
|
|
|
|
|
Zhu YG, Johnson TA, Su JQ, Qiao M, Guo GX, Stedtfeld RD, Tiedje JM (2013). Diverse and abundant antibiotic resistance genes in Chinese swine farms. Proceeding of the National Academy of Sciences 110(9):3435-3440.
Crossref
|
|