ABSTRACT
Poachers and bushmeat traders most of the time modify the wildlife materials before commercialization. Therefore, the morphological identification of species is no longer possible. However, accurate identification of the sale wildlife materials is essential for species identification, quantification and better conservation management. The aim of this study is to take advantages of specific gene sequences and in silico analysis to identify smoked and decapitated wildlife materials encountered on bushmeat markets and traditional restaurants in Center-Western Côte d’Ivoire. DNA from 352 bushmeat samples comprising 123 decapitated and smoked specimens was extracted, amplified by polymerase chain reaction (PCR) using the Cytb and COI primers and analyzed in silico. Up to 98.37% of the DNA from smoked bushmeat was successfully amplified. Assignment of Cytb and COI query sequences with reference sequences in the NCBI Genbank allowed to correct three misidentifications by traders. Thus, specimens identified as Heliosciurus sp, Phacochoerus aethiopicus africanus and Dendrohyrax dorsalis sylvestris by traders were assigned to Funisciurus sp, Tragelaphus scriptus, Crossarchus obscurus with 84.94, 100, 98.76% similarity respectively. Morphologically unidentifiable sample coded IP3CS1O was assigned to species Eidolon helvum with 99.53% similarity. The study shows the interest of DNA-based identification of bushmeat to circumvent misidentification for better management of wildlife biodiversity.
Key words: smoked bushmeat, misidentification, genetic typing, in silico analysis, identification, biodiversity, wildlife conservation.
Remote rural populations have always fed on wild fauna without really impinging on the conservation of biodiversity (Dufour et al., 2015). Today, the exploitation of wildlife products commonly known as bushmeat is much more for profit than for personal animal protein need (Chabi-Boni et al., 2019). The wildlife materials from the hunted animals are commercialized in different amorphous forms, like smoked meat, chunks decapitated, chopped meat, tanned skin, and so on (Kumar et al., 2014; Nishant et al., 2017). Due to the lack of morphological characters, identification of this wildlife material becomes inconclusive. Therefore, in order to recognize and accurately quantify poached wild animal for biodiversity and conservation purpose, it is important to use more sophisticated techniques which overcome morphological identification that has become impossible. Fortunately, advent of the use of genetic markers such as Cytochrome b, 12S ribosomal RNA, 16S ribosomal RNA, and Cytochrome c oxidase I are promising tools for species identification due to the bearing of nucleotide variation among the species (Hsieh et al., 2001; Panday et al., 2014; Kumar et al., 2019). In addition, millions of gene sequences are presently available online that permitted sequence similarity search to examine DNA data, generated from any biological material (Porter and Hajibabaei, 2018).
In Côte d'Ivoire, the district of Haut-Sassandra-Marahoué is rich in animal biodiversity in view of the existence of the national park of Marahoué and the protected reserves of Haut-Sassandra. However, with urban development, bushmeat preference by the urban populations and the evidence that the bushmeat trade is very lucrative, poachers are deeply attacking wildlife biodiversity (Yéboué et al., 2020). Thus, bushmeat is found on the markets and in restaurants in an amorphous way without identification possibility. However, it is more than essential to know the quantity of animals extracted from the bush to see the real intensity of biodiversity disturbance. The aim of the present study is to accurately identify smoked and decapitated bushmeat encountered on markets and traditional restaurants based on genetic markers and in silico analysis of DNA sequences.
Collection of wildlife sample
352 specimens of bushmeat comprising 123 smoked or decapitated specimens (Figure 1) were collected with traders or on restaurants for accurate identification purpose. Questions were addressed to traders and restaurant owners to have the first information about the identity of smoked or decapitated species.
DNA extraction, polymerase chain reaction (PCR) amplification and sequencing
Total genomic DNA extraction was performed using DNeasy® Tissue kit (Qiagen) according to manufacturer protocol and checked on 1% agarose gel for quality control. COI and Cytb primers were used (Table 1) to amplify partial mitochondrial DNA genes (Gaubert et al., 2014; Irwin et al., 1991; Kocher et al., 1989) PCR products representative of all species found according to field identification were coded and sent to BGI TECH SOLUTIONS (HONG-KONG) for sequencing.
DNA sequences assignment
DNA sequences provided in ABI format files were cleaned using Chromas Lite® 2.01 software to make them analyzable. The forward and reverse sequences of the genes were assembled and then corrected using DNA Baser Assembler 5.15.0 software and Chromas Lite® 2.01 software. In silico analysis of the sequences and phylogenetic characterization of the bushmeat species are made. Query sequences were aligned with reference sequences by submitting them to the public genomic database of the National Center for Biotechnology Information (NCBI) through its BLAST tool (Basic Local Alignment Search Tool; http://www.ncbi.nih.gov) (Altschul et al., 1990). The purpose of this alignment is to search areas of strong similarity between query and reference sequences existing in the public Genbank. For each alignment, identity percentage and a corresponding E-value are generated. The assignment selected is that of the coding sequence with the highest percentage of identity or similarity.
DNA amplification success rate
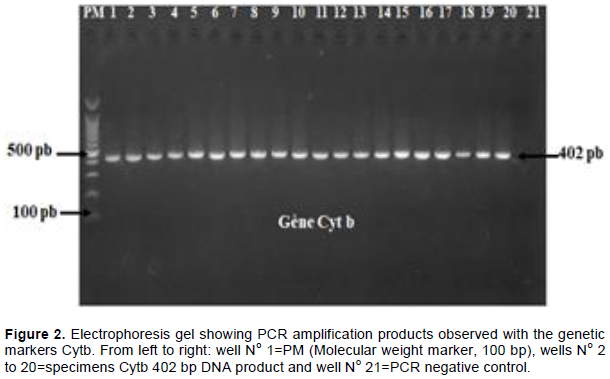
347 (98.58%) and 346 (98.30%) of the 352 specimens collected were successfully amplified respectively by Cytb and COI primers. Up to 98.37% (121/123) of the DNA from smoked bushmeat collected were successfully amplified (Figure 2).
Sequences assignment for species identification
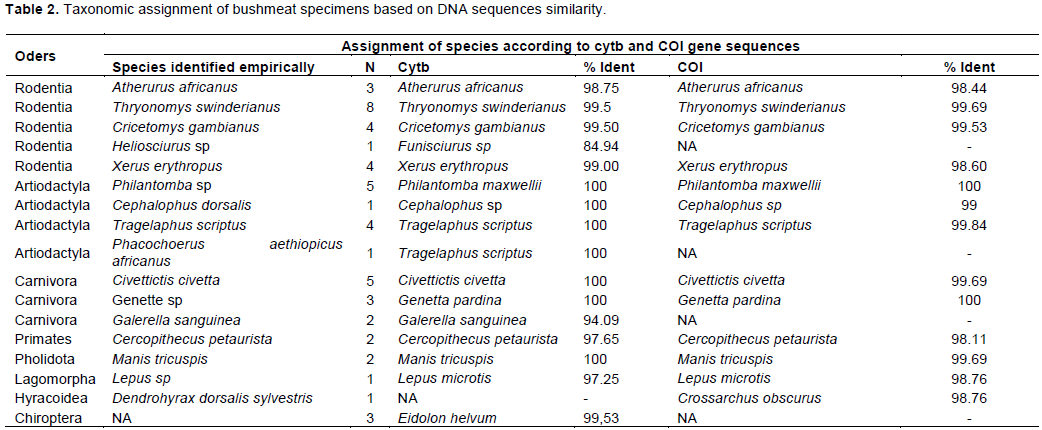
According to the information gave by the traders and our wildlife knowledge, the 50 representative specimens sent for sequencing are composed of sixteen species divided into eight orders: the order of Rodentia (05 species), Artiodactyla (04 species), Carnivora (03 species), Primates (01 species), Pholidota (01 species), Lagomorpha (01 species), Hyracoïdea (01 species), and Chiroptera (01 species). Alignment of Cytb and COI genes sequences of studied specimens, with reference sequences available in the NCBI Genbank revealed three misidentifications. Thus, the specimens morphologically identified as Heliosciurus sp was assigned to Funisciurus sp, with 84.94% similarity for the Cytb gene. The specimen of Artiodactyla code VP2PH1C, identified in the field as Phacochoerus aethiopicus africanus, was assigned to species Tragelaphus scriptus with 100% similarity with the Cytb gene (Table 2). The specimen initially identified as Dendrohyrax dorsalis sylvestris was assigned to Crossarchus obscurus with 98.76% similarity after alignment of its COI gene sequence. The sample coded IP3CS1O, collected in a smoked decapitated state that could not be identified morphologically was assigned to the species Eidolon helvum with 99.53% similarity after alignment of its Cytb gene sequence (Table 2). The remained species were accurately identified by traders according to DNA sequences assignment.
Taxonomic structure of poached species
The phylogenetic relationships of the specimens are described through dendrograms built according to the Neighbor-Joining model on the sequences of the Cytb and COI gene sequences generated. Analysis showed very narrow genetic distances between the specimens, defined in 15 taxonomic units: Civettictis civetta, Genetta pardina, Galerella sanguinea, Manis tricuspis, Cercopithecus petaurista, Eidolon helvum, Funisciurus sp, Atherurus africanus, Xerus erythropus, Philantomba maxwellii, Cephalophus dorsalis, Thryonomys swinderianus, Tragelaphus scriptus, Lepus microtis and Cricetomys gambianus. Specimen coded VP2PH1C, identified as Phacochoerus aethiopicus africanus, was found in the taxon Tragelaphus scriptus and shares a genetic similarity of 81% with specimen VP3G4R of this taxon (Figure 3).
Considering the COI gene, phylogenetic analysis of DNA sequences allowed the definition of 13 taxonomic groups comprising Philantomba maxwellii, Cephalophus dorsalis, Tragelaphus scriptus, Xerus erythropus, Cricetomys gambianus, Dendrohyrax arboreus, Atherurus africanus, Manis tricuspis, Civettictis civetta, Genetta pardina, Lepus microtis, Thryonomys swinderianus and Cercopithecus petaurista (Figure 4). Phylogenetic trees generated by the COI and Cytb genetic markers were in agreement in the assignment of the species observed except for the two species which could not be analyzed with the COI marker. A low bootstraps value is sometimes observed within the same taxonomic group due to intraspecific genetic diversity (Figures 3 and 4).
The precise identification of the wild species present on the bushmeat markets is a crucial concern for actors involved in the control and management of animal biodiversity (Ogden et al., 2009). Indeed, a large part of the bushmeat is sold in the form of carcasses or pieces of cut or smoked meat, which makes it difficult if not impossible to identify the species.
Underestimating the diversity of species sold in bushmeat markets in different forms results in underestimating the number and biomass of game species impacted by illegal hunting activity and trade. The possibility of identifying products of wild fauna via molecular genetic tools, with reference to global genomic databases (Ross et al., 2003; Ratnasingham and Hebert, 2007) is an innovation in the quantification of danger to wildlife in the reserves and national parks of Côte d'Ivoire.
In this study, molecular analysis using mitochondrial gene markers, COI and Cytb yielded an amplification success rate over 90% as predicted by Gaubert et al., (2014). One of the main steps in developing an accurate protocol for genetic analysis is choosing an appropriate area of ​​the genome for analysis, which will ensure a reliable diagnosis. In the mitochondrial genome of animal species, COI and Cyt b are more slowly evolving genes in terms of non-synonymous mutations, making them very suitable for the diagnosis of unique genetic signatures of species (Silva-Neto et al., 2016). In this study 15 species of mammals were discriminated with the Cyt b gene and 13 species with the COI gene. These results show that the Cytb gene is the most resolute marker. Similar studies have shown that the Cytb marker is the most efficient in molecular identification of mammalian species (Eaton et al., 2010; Olayemi et al., 2012; Naidu et al., 2012; Gaubert et al., 2014). However, the COI marker, in complementarity with Cytb, made it possible to identify certain species with a very high rate of similarity. Inaccuracies in species allocation have been observed in Cephalophus. Indeed, Cephalophus dorsalis identified morphologically was simply confirmed to belong to the genus Cephalophus. As mentioned by Munch et al. (2008) in their study, this lack of accuracy could be explained by the low representation of intraspecific diversity in Genbank. The discrepancies observed in this study could be due to several factors such as the insufficiency of genomic data on certain species in the public databases which should be updated and strengthened, the inaccurate identification of the species by the sellers and the smoked state. Molecular typing has proven to be very useful in resolving taxonomic confusion of smoked specimens, cryptic species and correcting misidentification in the field. The specimen VP2PH1C smoked and sold in our study area as Phacochoerius aethiopicus africanus was assigned after nucleotide sequence analysis to Tragelaphus scriptus (Bushbuck). These misidentifications find their justification in the cultural beliefs of the populations of west-central Côte d'Ivoire. Indeed, the Bushbuck is perceived as a prohibited species for these populations because it would be the preferred species for mystical practices. Thus, restaurants, to increase their income margin, sell their cargo at the price of other game. This assertion is supported by the work of Gaubert et al., (2014). These authors in their study suspected that traders increase their income by selling domestic animals (pigs) at the cost of wild game to Muslims in Southwestern Nigeria. Other discrepancies between morphological and molecular identification were also observed in this study with smoked specimens difficult to distinguish with the naked eye from closely related species. This is the case of Heliosciurus sp morphologically identified and Funisciurus sp attributed by mtDNA. This result could be explained by natural hybridization between species, incomplete sorting of lines or poor delineation of boundaries between species (Gongora et al., 2011). This study revealed another case of discrepancy; that of a morphologically unidentified specimen assigned to the species Eidolon helvum by Cyt b DNA marker. The same event was highlighted by the study by Olayemi et al., (2012) in Cricetomys gambianus with the Cytb gene. Phylogenetic analysis of the COI and Cytb gene sequences successfully grouped the collected bushmeat specimens into target taxa. In addition, genetic variability was observed within the taxa obtained. This variability is believed to be due to the probable origin of many taxa that formed the targets of this study or to the hybridization of sister taxa. These results are in agreement with those of Ekrem et al. (2007) who experienced non-monophyllous taxa by phylogenetic analysis.
The study of poached and smoked wild animals in the central-west of Côte d'Ivoire aimed to identify species, morphologically unidentifiable, found in bushmeat markets and restaurants by genetic typing in order to validate these molecular tools in quantifying the loss of biodiversity linked to poaching in Côte d’Ivoire. Over 90% amplification success was observed with the COI and Cytb markers. The smoky condition of the specimens did not influence the amplification in any way. This study shows that the Cytb gene is the best marker for molecular identification of mammalian species. However, the COI marker, in complementarity with Cyt b, makes it possible to identify certain species at a very high rate of similarity, thus giving the opportunity to correct certain discrepancies in the identification of the species.
The authors have not declared any conflict of interests.
REFERENCES
|
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990). Basic local alignment search tool. Journal of Molecular Biology 215(3):403-410.
Crossref
|
|
|
|
Chabi-Boni SD, Natta AK, Nago SGA, Mensah GA (2019). Diversité des Espèces de Faunes Chassées et Impact sur la Biodiversité Animale (Nord-Ouest du Bénin). European Scientific Journal 15:1857-7881.
Crossref
|
|
|
|
|
Dufour S, Béné JCK, Dao D, N'guessan E (2015). Gestion durable de la faune et des ressources cynégétiques en Côte d'Ivoire. Rapport pour les états généraux de la forêt, de la faune et des ressources en eau, 101 p.
|
|
|
|
|
Eaton MJ, Meyers GL, Kolokotronis SO, Leslie MS, Martin AP, Amato G (2010). Barcoding bushmeat: molecular identification of Central African and South American harvested vertebrates. Conservation Genetics 11(4):1389-1404.
Crossref
|
|
|
|
|
Ekrem T, Willassen E, Stur E (2007). A comprehensive DNA sequence library is essential for identification with DNA barcodes. Molecular Phylogenetics and Evolution 43(2):530-542.
Crossref
|
|
|
|
|
Gaubert P, Njiokou F, Olayemi A, Pagani P, Dufour S, Danquah E, Nutsuakor MEK, Ngua G, Missoup AD, Tedesco PA, Dernat R, Antunes A (2014). Bushmeat genetics: setting up a reference framework for the DNA typing of African forest bushmeat. Molecular Ecology Resources 15(3):633-651.
Crossref
|
|
|
|
|
Gongora J, Cuddahee RE, Nascimento FFD, Palgrave CJ, Lowden S, Ho SY, Simond D, Damayanti CS, White DJ, Tay WT, Klingel H, Rodrigues-Zarate CJ, Allen K, Moran C, Randi E (2011). Rethinking the evolution of extant subâ€Saharan African suids (Suidae, Artiodactyla). Zoologica Scripta 40(4):327-335.
Crossref
|
|
|
|
|
Hsieh HM, Chiang HL, Tsai LC, Lai SY, Huang NE, Linacre A, Lee JC (2001). Cytochrome b gene for species identification of the conservation animals. Forensic Science International 122:7-18.
Crossref
|
|
|
|
|
Irwin DM, Kocher TD, Wilson AC (1991). Evolution of the cytochromeb gene of mammals. Journal of Molecular Evolution 32(2):128-144.
Crossref
|
|
|
|
|
Kocher TD, Thomas WK, Meyer A, Edwards SV, Pààbo S, Villablanca FX, Wilson AC (1989). Dynamics of mitochondrial DNA evolution in animals: Amplification and sequencing with conserved primers. Proceedings of the National Academy of Sciences USA 86:6196-6200.
Crossref
|
|
|
|
|
Kumar VP, Kumar D, Goyal SP (2014). Wildlife DNA forensic in curbing illegal wildlife trade: species identification from seizuresInternational Journal of Forensic Science and Pathology 2:38-42.
Crossref
|
|
|
|
|
Kumar V, Chandraa K, Kundua S, Tyagia K, Laskarb AB, Singhaa D, Chakrabortya R, Pakrashia A (2019). Utility of mitochondrial DNA in wildlife forensic science: reliable identification of confiscated materials from Eastern India. Mitochondrial DNA part b, 4(1):583-588.
Crossref
|
|
|
|
|
Munch K, Boomsma W, Huelsenbeck JP, Willerslev E, Nielsen R (2008). Statistical assignment of DNA sequences using Bayesian phylogenetics. Systematic Biology 57:750-757.
Crossref
|
|
|
|
|
Naidu A, Fitak RR, Munguia-Vega A, Culver M (2012). Novel primers for complete mitochondrial cytochrome b gene sequencing in mammals. Molecular Ecology Resources 12:191-196.
Crossref
|
|
|
|
|
Nishant K, Vrijesh KY, Ajay KR (2017). Wildlife forensic: current techniques and their limitations. Journal of Forensic Science and Criminology 5(4):402 p.
Crossref
|
|
|
|
|
Ogden R, Dawnay N, McEwing R (2009). Wildlife DNA forensics-bridging the gap between conservation genetics and law enforcement. Endangered Species Research 9:179-195.
Crossref
|
|
|
|
|
Olayemi A, Nicolas V, Hulselmans JAN, Missoup AD, Fichet-Calvet E, Amundala D, Dudu A, Dierckx T, Wendelen W, Leirs H, Verheyen E (2012). Taxonomy of the African giant pouched rats (Nesomyidae: Cricetomys): molecular and craniometric evidence support an unexpected high species diversity. Zoological Journal of the Linnean Society 165(3): 700-719.
Crossref
|
|
|
|
|
Panday R, Jha DK, Thapa N, Pokharel BR, Aryal NK (2014). Forensic wildlife parts and their product identification and individualization using DNA barcoding. The Open Forensic Science Journal 7:6-13.
Crossref
|
|
|
|
|
Porter TM, Hajibabaei M (2018). Over 2.5 million COI sequences in GenBank and growing. PLoS ONE 13(9):e0200177.
Crossref
|
|
|
|
|
Ratnasingham S, Hebert PDN (2007). BOLD: The barcode of life data system. Molecular Ecology 7:355-364.
Crossref
|
|
|
|
|
Ross HA, Lento GM, Dalebout ML, Goode M, Ewing G, McLaren P, Rodrigo AG, Lavery S, Baker CS (2003). DNA surveillance: web-based molecular identification of whales, dolphins, and porpoises. Journal of Heredity 94(2):111-114.
Crossref
|
|
|
|
|
Silva-Neto AA, Ferreira PB, Torres RA, Texeira RHF, Duarte JMB, Barbosa AC, Vargase RC, Garcia JE (2016). Diagnostic Cytochrome b gene profiles for the identification of paca (Cuniculus paca) bushmeat: implications for the monitoring of illegal hunting and wildlife trade. Brazilian Journal of Biology 76(1):55-58.
Crossref
|
|
|
|
|
Yéboué KF, Koffi M, Sylla I., Abé AI, Ahouty B, N'djetchi KM, Simaro S, Konan T, Tidou SA, Béné JCK (2020). Quantifying poached wildlife mammal species in Center-western region of Côte d'Ivoire. Journal of Ecology and the Natural Environment 12(3):120-128.
Crossref
|
|