ABSTRACT
Production of groundnuts (Arachis hypogaea L.) in Western Kenya is mainly constrained by groundnut rosette disease (GRD) which cause up to 100% yield loss. This disease expresses different symptoms as a result of variations in the groundnut rosette virus (GRV) associated satellite-ribonucleic acid (GRV Sat-RNA). Over the past 20 years, no work had been done to document the status of the disease in Kenya. Additionally, no sequences of any of the GRD associated viruses were available in the GeneBank from Kenya. This study determined the distribution of GRD and the genetic diversity of GRV Sat-RNA. Sampling was done in main groundnut growing areas of Western Kenya during the long and short rain seasons in 2016/2017. Total RNA was extracted from the leafy samples collected using RNeasy Mini Kit (Qiagen) according to the manufacturers’ protocol and used for double stranded cDNA synthesis using the SuperScript II kit. DNA libraries were sequenced on the Illumina MiSeq platform (Illumina). Reads were used for de novo assembly and contigs aligned to the viral genomes database using CLC Genomics Workbench 10.1.2. The assembled contigs were subjected to a BLASTn search against the GenBank database. Average GRD incidence was 53 and 41% in the short and long rain seasons, respectively. Chlorotic rosette was the dominant symptom followed by green rosette and mosaic. Nucleotide sequences of Sat-RNA revealed identities of 88 to 100% between the Kenyan isolates and those from Malawi, Nigeria and Ghana. All Kenya isolates clustered closest with green rosette variants of Malawi except one which clustered with chlorotic/yellow blotch variants. Rosette is widely distributed in Western Kenya and occurs wherever groundnuts are grown. The variations of GRD symptoms in Western Kenya could be due to the existence of different variants of Sat-RNA or other agents.
Key words: Groundnuts, satellite-ribonucleic acid (Sat-RNA), diversity, Western Kenya.
Groundnut (Arachis hypogaea L.) is the fifth most important annual oilseed and food legume crop. It is grown in diverse environments throughout the semi-arid and sub-tropical regions, in nearly 100 countries, in the six continents of the world (Kumar and Waliyar, 2007). Groundnut production is of great value in terms of income and nutrition for smallholder farmers in East Africa (Kidula et al., 2010; Okello et al., 2010). Resource poor smallholder farmers grow nearly 75 to 80% of the world’s groundnuts in developing countries obtaining yields of 500 to 800 kg/ha, as opposed to the potential yield of >2.5 t/ha (Kayondo et al., 2014). In Western Kenya, an average of 600 to 700 kg/ha is achieved which is less than 30 to 50% of the potential yield (Kidula et al., 2010). Low yields are mainly attributed to poor quality seeds, drought, poor agronomic practices, numerous pests and diseases caused by numerous pathogenic viruses, fungi, bacteria and nematodes (Mutegi, 2010; Okello et al., 2010). Among the viral diseases, groundnut rosette disease (GRD) is the most devastating in sub-Saharan Africa (SSA) causing an estimated annual loss of US$156 million every year (Waliyar et al., 2007). The disease is caused by association between groundnut rosette assistor virus (GRAV), groundnut rosette virus (GRV) and a Satellite-RNA (Sat-RNA) of GRV (Taliansky and Robinson, 2003). Variations in Sat-RNA have been shown to be responsible for different rosette symptoms (chlorotic, green and mosaic rosette) (Taliansky and Robinson, 1997; Olorunju et al., 2001; Kayondo et al., 2014). Both chlorotic and green rosette symptoms occur throughout the SSA, and sometimes occur in the same field (Mugisa et al., 2016). A less common third symptom variant, called mosaic rosette, resulting from mixed infection by the Sat-RNA causing chlorotic and green mottled variant, has been reported from East Africa (Scott et al., 1996; Waliyar et al., 2007). Infected groundnut leaves may also show symptoms other than the typical chlorotic or green rosette (Naidu et al., 1999).
In Eastern Uganda, green rosette symptoms predominate (Okello et al., 2014). This is in contrast with the findings of Wangai et al. (2001), who reported that chlorotic rosette symptoms of GRD have been the predominant form throughout SSA and Western Kenya. The dynamics of the GRD virus symptomatology, therefore, needs constant monitoring. For example, in Nigeria, a there was shift from green to chlorotic rosette over a period of about 20 years. The shift could be due to changes in the genome sequences of GRD associated agents or other factors (Okello et al., 2014).
Survey done by Wangai et al. (2001) showed that GRD incidence ranged between 40% in areas of Western Kenya surveyed in the groundnut growing seasons of 1997-1998 and Sat-RNA shared 89 to 95% nucleotide identity with those from Malawi and Nigeria. Since then, no other survey has been conducted to ascertain the current status of GRD in the region. In addition, no genomic sequences of any of the GRD associated viruses existed in the GeneBank from Western Kenya. With the dynamics of the disease, this hinders proper diagnosis of GRD and development of management strategies. This study determined the distribution of GRD and assessed the sequence diversity of Sat-RNA of isolates from Western Kenya.
Field survey
The GRD diagnostic sampling was conducted in all the major groundnut growing areas in Western Kenya during the short rains (October to December 2016) and long rains season (May to July, 2017). The following counties were covered: Bungoma, Busia, Homabay, Kakamega, Siaya and Vihiga. Sampling of groundnut farms was done by stopping at regular predetermined intervals, of 3 to 8 km along motorable roads that traverses each sampling area. The survey was conducted, by walking through groundnut field and visually inspecting groundnut crops for symptomatic leaves. Disease incidence was calculated according to Reddy (1991), as the percentage of plants showing GRD virus symptoms, to the total number of plants observed in the field. GRD viral incidence was scored using a rating scale according to Reddy (1991) where: low incidence = 1-20%; moderate incidence = 21-49%, and high incidence = 50-100%. The types of GRD symptoms observed were recorded. The collected data on GRD virus incidence and severity, was subjected to analysis of variance (ANOVA), using Statistical Analysis System (SAS) program version 9.3.1 software (SAS Institute, 2013). Pairwise comparisons of means were done using Least Significance Differences (LSD) for multiple-means comparison method at P ≤ 0.05 confidence level.
Symptomatic and asymptomatic leaves were collected in 10 ml falcon tubes containing RNAlater® RNA Stabilization Solution and put in a cool box. The samples were kept in the fridge and used for molecular studies. Geographical Positioning System (GPS) (entrex venture HC GARMINTM), was used to record the latitude, longitude and altitude of the sampled farms.
RNA extraction, sequencing and sequence analyses
Total RNA was extracted from the leaf samples using RNeasy Mini Kit (Qiagen) according to the manufacturers’ protocol and used for double-stranded cDNA synthesis using the SuperScript II (Thermo Fisher Scientific, Waltham, USA) kit. The cDNA was column-purified with the DNA Clean and ConcentratorTM-5-DNA kit (Zymo Research, Irvine, USA). The samples were then processed with the transposon-based chemistry library preparation kit (Nextera XT, Illumina) following manufacturer’s instructions. The fragment sizes structure of the DNA libraries was assessed using the Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, USA). The indexed denatured DNA libraries were sequenced (200-bp paired-end sequencing) on the Illumina MiSeq platform (Illumina).
Reads quality check was done using FastQC (version 0.11.5). Reads were then trimmed to remove poor quality sequences using Trimmomatic (V 0.36) software (Bolger et al., 2014). Trimmed reads (Haas et al., 2013) were used for de novo assembly and contigs aligned to the viral genomes database (ftp://ftp. ncbi.nih.gov/genomes/Viruses/all.fna.tar.gz/, downloaded on October 2017) using CLC Genomics Workbench 10.1.2. The assembled contigs were subjected to a BLASTn search against the GenBank database (Altschul et al., 1990). Complete and partial GRV Sat-RNA sequences used for comparison and phylogenetic analyses were retrieved from GenBank (http://www.ncbi.nlm.nih.gov/). Phylogenetic analyses and comparisons were performed using the MEGA X (Kumar et al., 2018). Tobacco bushy top virus - KU997687.1 TBTV was used as an outgroup.
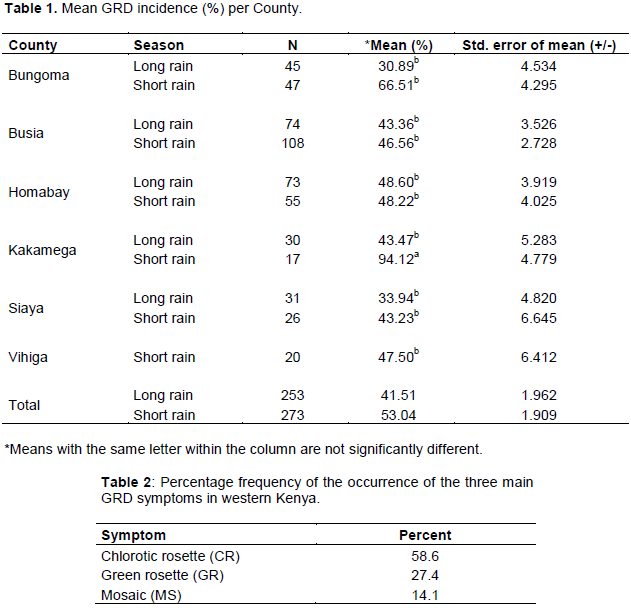
A total of 526 farms were sampled in six (6) counties (253 in long rain and 273 in short rain). Generally, GRD incidence was high during the short rain season (53%) than the long rain season (41%) in all counties. High mean GRD incidence was recorded in Kakamega in the short rain season (68.92%) while moderate incidence was in Bungoma (30.89%) during the long rain season. There was a significant difference in GRD incidence among the counties (p=0.011, df=521, F=3.322). Siaya County had the overall lowest incidence which was significantly different from that of Kakamega but did not vary significantly from that of Bungoma, Busia, Vihiga and Homabay counties (Table 1).
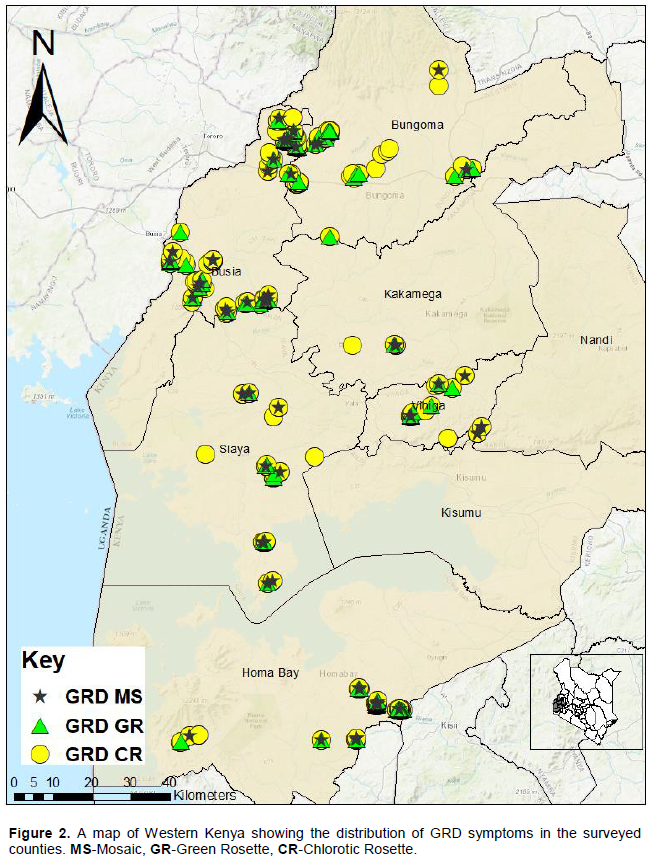
Generally, GRD infected plants were dwarf with increased tillering although some were tall but expressed other major symptoms associated with GRD. The main symptoms observed in all counties in order of abundance, starting from the most common, were chlorotic rosette, green rosette and mosaic. Chlorotic rosette was recorded in 58.6% of the surveyed farms, green rosette in 27.4% while mosaic was observed 14.1% of farms (Table 2). Other symptoms observed were leaf rolling, upward leaf curling and severe leaf bunching (Figure 1). The distribution of the major symptoms is shown in Figure 2.
Diversity of GRV Sat-RNA
Six complete genomes of GRV Sat-RNA were assembled. The sequences varied slightly in the number of nucleotides (nt) ranging between 896 and 901 nt (Table 3).
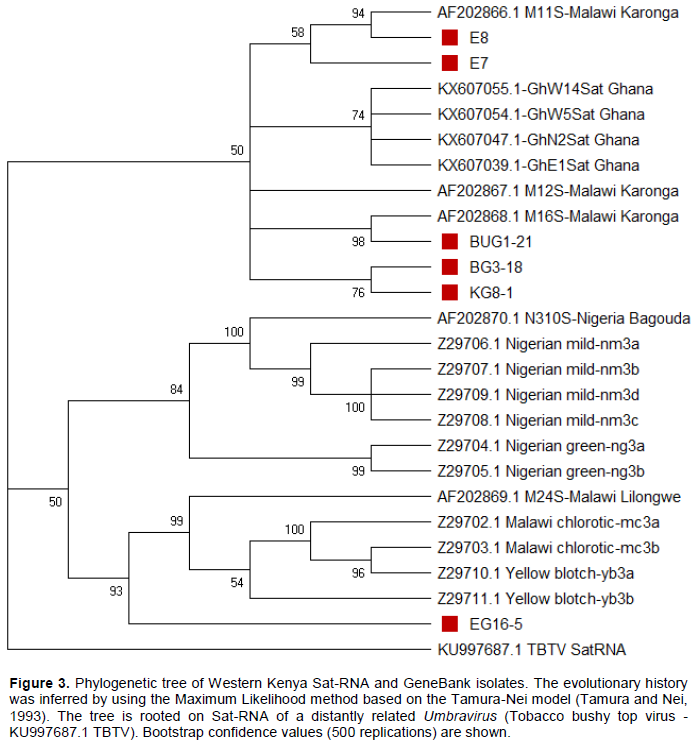
The six Sat-RNAs from Kenya were then compared with those from the GeneBank. In the phylogenetic tree, all Kenyan isolates formed two distinct clusters together with Malawian isolates. Isolates E7 and E8 clustered with M11S, isolates BUG1-21, BG3-18 and KG8-1 clustered together with M16S while isolate EG16-5 is grouped with M24S. All Nigerian isolates clustered together is similar to Ghanaian isolates. Sequence identities between 88 and 100% of the Kenyan isolates and those from Malawi, Nigeria and Ghana were revealed. Very close identities between 92 and 100% were observed between the Kenyan isolates and those from Malawi, followed by Nigerian isolates (90-93%) and least with Ghanaian isolates (86-89%). Isolate BUG1-21 had 100, 99 and 98% identities with M16S, M12S, and M11S, respectively, which are all green rosette variants and 94% with M24S (chlorotic variant). While the other Western Kenya isolates (KG8-1, BUG1-21, BG3-18, E7 and E8) had 92 to 95% identity with Malawian isolate M24S (chlorotic rosette variant), isolate EG16-5 (Kakamega) showed the closest identity (97%) with this isolate. The same isolate EG16-5 was the only that clustered together with M24S, all chlorotic isolates (Z29702.1, Z29703.1) and yellow blotch (Z29710.1, Z29711.1). Isolates E7 and E8 were closest to Malawian isolate M11S with 97 and 99% identity, respectively. Isolates BG3-18 and KG8-1 were closest to Malawian isolates M16S displaying 97% identity (Figure 3).

Groundnut rosette disease is the most prevalent disease of groundnuts in Western Kenya. The disease was recorded in every county that was surveyed with incidences of up to 100%. The short rain season recorded higher incidence (53%) than the long rains (41%). This can be attributed to the high vector pressure during the short rains as compared to the long rains season when the aphid pressure is low as a result of heavy rains that wash the insects away. This concurs with the findings by Mugisa et al. (2016) that periods of long rains negatively affected GRD progression when aphid pressure is low.
All major GRD symptoms were observed in the surveyed region with chlorotic rosette being the most prevalent followed by green rosette. This concurs with Wangai et al. (2001) who reported chlorotic rosette to be the most prevalent GRD symptom in the region. The high prevalence of the chlorotic rosette could also be attributed to its higher transmission efficiency compared to green rosette. In a study, Misari et al. (1988a), reported minimum acquisition feeding periods of 4 and 8 h for chlorotic and green rosette, respectively and the median latent periods of 26.4 and 38.4 h, respectively, for chlorotic and green rosette. In this study, a new symptom, the mosaic, which had not been previously reported in Western Kenya, was observed in most of the surveyed counties. This suggests that there is evolution of new variants of Sat-RNA that might be causing these new symptoms. A total of 10 variants of Sat-RNA have been reported as being associated with the various GRD symptoms (Blok et al., 1994). A mixture of either variants, especially the chlorotic and green rosette and/or the mild ones, are likely to induce the mosaic symptoms (Naidu et al., 1998). It is therefore possible that the variants of sat-RNA reported in this study occur in Western Kenya in mixed infections, thus causing the mosaic observed. It is worth noting that from the Next Generation Sequences (NGS) used in this study, order than GRV Sat-RNA, other viruses were detected (data not shown) and could be the reason for some of the new symptoms observed on groundnuts (Mukoye et al., 2018).
The Western Kenya Sat-RNAs sequences showed close identity (92-100%) to Malawian isolates than those from Ghana and Nigeria (88-93%). This implies that the genetic diversity of the Sat-RNA become more varied with wide geographical distance. Kenya and Malawi are located in Eastern Africa while Ghana and Nigeria are in West Africa thus having a wider geographical separation than Malawi. This finding is in line with Wangai et al. (2001) who observed a closer sequence relationship between Kenyan Sat-RNA isolates and those from Malawi. However, this study has reported sequence identity of up to 100% with Malawian isolates as opposed to 95% reported by Wangai et al. (2001). This suggests that more variants of Sat-RNA exist in Western Kenya that are contributing to the diverse symptoms expressed by GRD. Since this study used NGS which has been demonstrated to be more reliable in detection of new or poorly characterized viruses (Rott et al., 2017), it has revealed new variants of Sat-RNA in Western Kenya. Besides, there were variations among the Western Kenya Sat-RNA isolates similar to Malawian isolates where they formed distinct clusters in the phylogenetic tree. The isolate EG16-5 was the most distinct and clustered together with chlorotic and yellow blotch Sat-RNA variants. This suggests that this isolate is related to the chlorotic rosette symptom that was prevalent in the surveyed areas.
This study concludes that GRD is still the major viral disease of groundnuts in Western Kenya and occurs wherever groundnuts are grown in the region. The disease expresses varied symptoms with chlorotic rosette being the most prevalent form. The observed variations in the symptoms were due to the presence of diverse variants of the symptom inducing agent, Sat-RNA. The use of NGS has revealed that new variants of Sat-RNA exist in Western Kenya.
The six Kenyan Sat-RNAs have been deposited in the GeneBank with accession numbers LC469779, LC472299, LC472300, LC472301, LC472302 and LC472303.
The authors have not declared any conflict of interests.
This work was funded by Royal Society of UK, International Foundation for Science, National Research Fund (NRF-Kenya) and ILRI BecA hub. They are grateful to Dr. Wellington Ekaya and the entire BecA capacity building team for allowing this work to be done at the BecA labs.
REFERENCES
|
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990). Basic local alignment search tool. Journal of Molecular Biology 215:403-410.
Crossref
|
|
|
|
Blok VC, Ziegler A, Robinson DJ, Murant AF (1994). Sequences of 10 variants of the satelitte- like RNA -3 of groundnut rosette virus. Virology 202:25-32.
Crossref
|
|
|
|
|
Bolger AM, Lohse M, Usadel B (2014). Trimmomatic: A flexible trimmer for Illumina Sequence Data. Bioinformatics, btu170.
Crossref
|
|
|
|
|
Haas BJ, Papanicolaou A, Yassour M, Grabherr M, Blood PD, Bowden J, Couger MB, Eccles D, Li B, Lieber M, MacManes MD, Ott M, Orvis J, Pochet N, Strozzi F, Weeks N, Westerman R, William T, Dewey CN, Henschel R, LeDuc RD, Friedman N, Regev A (2013). De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nature Protocols 8:1494-1512.
Crossref
|
|
|
|
|
Kayondo SI, Rubaihayo PR, Ntare BR, Gibson PI, Edema R, Ozimati A, Okello DK (2014). Genetics of resistance to groundnut rosette virus disease: African Crop Science Journal 22:21-29.
|
|
|
|
|
Kidula N, Okoko N, Bravo-Ureta BE, Thuo M, Wasilwa L (2010). A preliminary analysis of yield differences in groundnuts between research and non-research farmers in Kenya. In paper presented at the 12th KARI biennial scientific conference, 8-12 November 2010, Naiobi Kenya.
|
|
|
|
|
Kumar S, Stecher G, Li M, Knyaz C, Tamura K (2018). MEGA X: Molecular Evolutionary Genetics Analysis across computing platforms. Molecular Biology and Evolution 35:1547-1549.
Crossref
|
|
|
|
|
Kumar PL, Waliyar F (2007). Diagnosis and detection of viruses infecting ICRISAT mandate crops: Methods Manual. Patancheru 502 324, Andhra Pradesh, India; International Crops Research Institute for the Semi-Arid Tropics.133 p.
|
|
|
|
|
Misari SM, Abraham JM, Demski JW, Ansa OA, Kuhn CW, Casper R, Breyel E (1988a). Aphid transmission of the viruses causing chlorotic and green rosette diseases of peanut in Nigeria. Plant Disease 72:250-253.
Crossref
|
|
|
|
|
Mugisa IO, Karungi J, Akello B, Ochwo-Ssemakula MKN, Biruma M, Okello DK, Otim G (2016). Determinants of groundnut rosette virus disease occurrence in Uganda. Elseviercropprotectionjournal.
Crossref
|
|
|
|
|
Mukoye B, Mangeni BC, Sue J, Ndong'a MFO, Were HK (2018). Next generation sequencing as a tool in modern pest risk analysis: a case study of groundnuts (Arachis hypogaea) as a potential host of new viruses in Western Kenya. Kenya Plant Health Inspectorate Service 2nd phytosanitary conference, 4th - 8th June, 2018, Nairobi - Kenya.
|
|
|
|
|
Mutegi CK (2010). The extend of aflatoxin and aspergillus section flavi, penicillium spp. and Rhizopus spp. contamination of peanuts from households in Western Kenya and the causative factors of contamination. PhD dissertation, University of Kwazulu-Natal,Pietermaritzburg . South Africa.
|
|
|
|
|
Naidu RA, Bottenberg H, Subrahmanyam P, Kimminns FM, Robinson DJ, Thresh JM (1998). Epidemiology of groundnut rosette virus disease: Current status and future research needs. Annals of Applied Biology 132:525-548.
Crossref
|
|
|
|
|
Naidu RA, Kimmins FM, Deom CM, Subrahimanyam P, Chiyeubekeza AJ, van der Merwe PJA (1999). Groundnut rosette: Avirus disease affecting groundnut production in Sub-Saharan Africa. Plant Disease 83:700-709.
Crossref
|
|
|
|
|
Okello DK, Birima M, Deom CM (2010). Overview of groundnut research in Uganda: Post, present and future. African Journal of Biotechnology 9:6448-6459.
|
|
|
|
|
Okello DV, Akello LB, Tukamuhabwa P, Odongo TL, Ochwo-Ssemakula M, Adriko J, Deom CM (2014). Groundnut rosette disease symptom types, distribution and management of the disease in Uganda. African Journal of Plant Science 8:153-163.
|
|
|
|
|
Olorunju PE, Ntare BR, Pande S, Reddy SV (2001). Additional sources of resistance to groundnut rosette disease in groundnut germplasm and breeding lines. Annals of Applied Biology 159:259-268.
Crossref
|
|
|
|
|
Reddy DVR (1991). Groundnut viruses and virus diseases; Distribution, identification and control. Review of Plant Pathology 70:665-678.
|
|
|
|
|
Rott M, Xiang Y, Boyes I, Belton M, Saeed H, Kesanakurti P, Hayes S, Lawrence T, Birch C, Bhagwat B, Rast H (2017). Application of Next Generation Sequencing for diagnostic testing of tree fruit viruses and viroids. Plant Disease 101:1489-1499.
Crossref
|
|
|
|
|
SAS Institute (2013). SAS/STAT 9.3.1: User guide. SAS Publishers, India.
|
|
|
|
|
Scott KP, Farmer MJ, Robinson DJ, Torrence L, Murant AF (1996). Comparison of the coat protein of groundnut rosette assistor virus with those of other luteovirus. Annals of Applied Biology 128:77-83.
Crossref
|
|
|
|
|
Taliansky ME, Robinson DJ (1997). Trans-acting untranslated elements of groundnut rosette virus satelitte RNA are involved in symptom production. Journal of General Virology 78:1277-1285.
Crossref
|
|
|
|
|
Taliansky ME, Robinson DJ (2003). Molecular Biology of umbraviruses: Phantom warriors. Journal of General Virology 84:1951-1960.
Crossref
|
|
|
|
|
Tamura K, Nei M (1993). Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Molecular Biology and Evolution 10:512-526.
|
|
|
|
|
Waliyar F, Kumar PL, Ntare BR, Monyo E, Nigam SN, Reddy AS, Osiru M, Diallo AT (2007). A Century of Research on Groundnut Rosette Disease and its Management. Information Bulletin no.75.Patancheru 502 324, Andhra Pradesh, India. International Crops Research Institute for the Semi-Arid Tropics 40 p. ISBN 978-92-9066-501-4.
|
|
|
|
|
Wangai AW, Pappu SS, Pappu HR, Okoko N, Deom CM, Naidu RA (2001). Distribution and characteristics of groundnut rosette disease in Kenya. Plant Disease 85:470-474.
Crossref
|
|