Full Length Research Paper
ABSTRACT
INTRODUCTION
MATERIALS AND METHODS
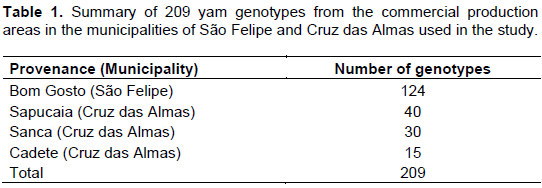
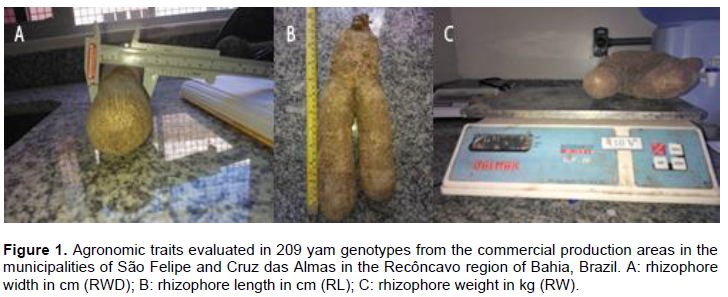
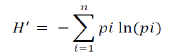
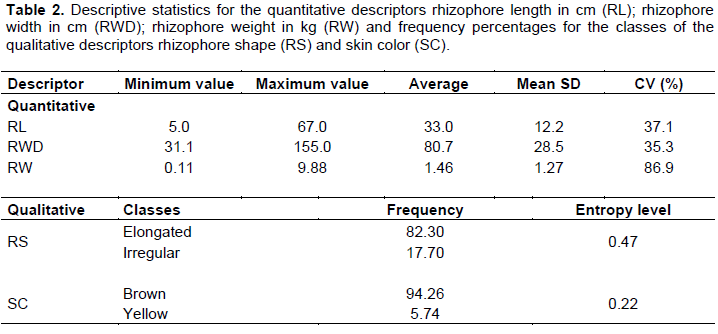
RESULTS AND DISCUSSION
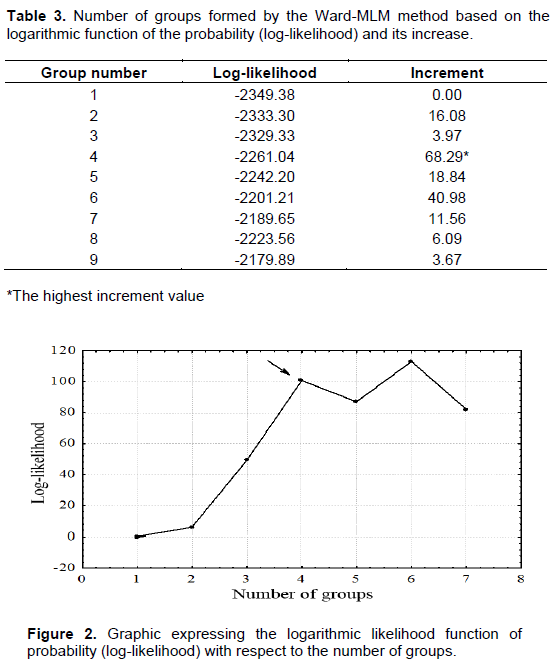
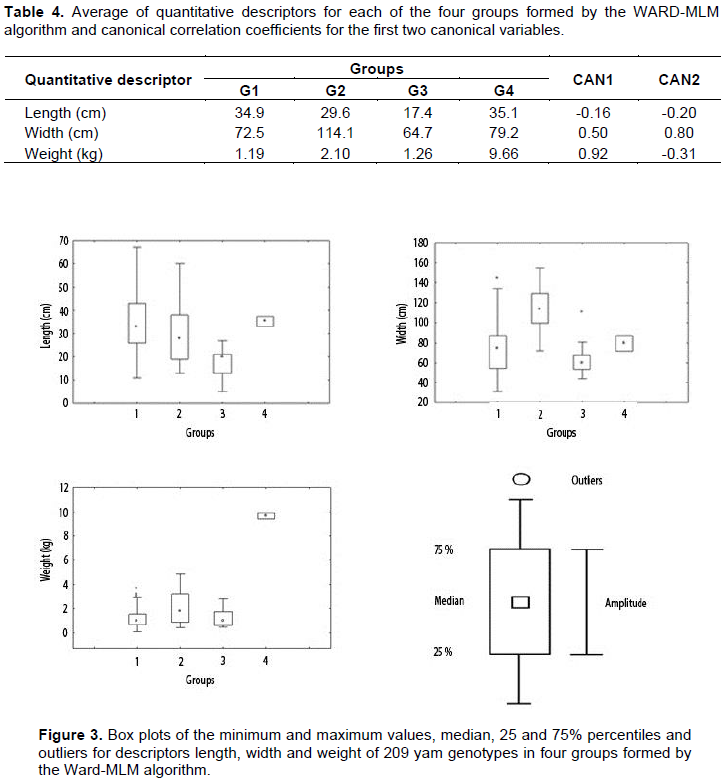
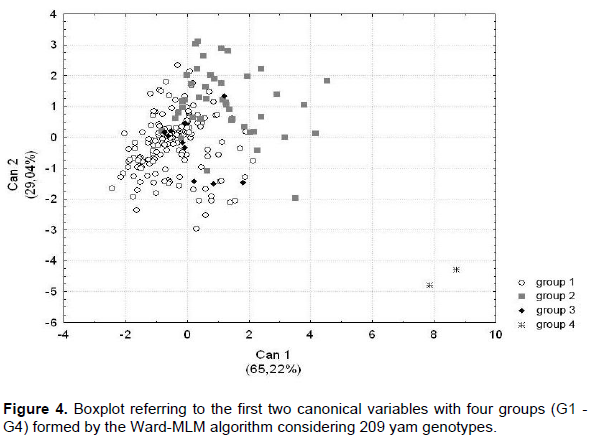
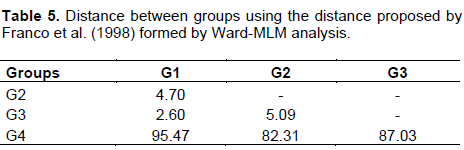
CONCLUSIONS
CONFLICT OF INTERESTS
REFERENCES
|
Arnau G, Bhattacharjee R, MN S, Chair H, Malapa R, Lebot VKA, Perrier X, Petro D, Penet L, Pavis C (2017). Understanding the genetic diversity and population structure of yam (Dioscorea alata L.) using microsatellite markers. PLoS one 12(3):e0174150. |
|
|
Asare S, Tetteh AY, Twumasi P, Adade KB, Akromah RA (2016). Genetic diversity in lowland, midaltitude and highland African maize landraces by morphological trait evaluation Afr. J. Plant Sci. 10(11):246-257. |
|
|
Barbé TC, Amaral Júnior AT, Gonçalves LSA, Rodrigues R, Scapim CA (2010). Association between advances generations and genealogy in inbred lines of snap bean by the Ward-Modified Location Model. Euphytica 173:337-343. |
|
|
Cabral PDS, Soares TCB, Gonçalves LSA, Amaral Júnior AT, Lima ABP, Rodrigues R, Matta FP (2010). Quantification of the diversity among common bean accessions using Ward-MLM strategy. Pesqui. Agropecu. Bras. 45(10):1124-1132. |
|
|
Carvalho PCL, Teixeira CA, Borges AJ (2009). Diversidade genética em Dioscorea spp. no Recôncavo da Bahia. Rev. Bras. Agroecol. 4(2):515-519. |
|
|
Cole-Rodgers P, Smith DW, Bosland PW (1997). A novel statistical approach to analyze genetic resource evaluations using Capsicum as an example. Crop Sci. 37:1000-1002. |
|
|
Cruz CD (2013). GENES - a software package for analysis in experimental statistics and quantitative genetics. Acta Sci. Agron. 35(3):271-276. |
|
|
Da Silva Dantas DF, De Oliveira AP, Dantas TAG, Da Silva Bandeira N, Neto ADG (2013). Produção do inhame adubado com doses de N e esterco bovino. Semin: Cienc. Agrar. 34:3389-3396. |
|
|
Dansi A, Dantsey-Barry H, Dossou-Aminon I, Kpenu EK, Agré AP, Sunu YD, Kombate K, Loko YL, Dansi M, Assogba P, Voudohe R (2013). Varietal diversity and genetic erosion of cultivated yams (Dioscorea cayenensis Poir-D. rotundata Lam complex and D. alata L.) in Togo. Int. J. Biodivers. Conserv. 5(4):223-239. |
|
|
FAOSTAT (2014). Food and agriculture organisation of the United Nation. http://www.fao.org/faostat/en/#data/QC. Accessed on 12.01.2016. |
|
|
Franco J, Crossa J, Villase-or J, Taba S, Eberhart SA (1998). Classifyng genetic resources by categorical and continuous variables. Crop Sci. 38:1688-1696. |
|
|
Gonçalves LSA, Rodrigues R, Amaral Júnior AT, Karasawa M, Sudré CP (2009). Heirloom tomato gene bank: assessing genetic divergence based on morphological, agronomic and molecular data using Ward-modified location model. Genet. Mol. Res. 8:364-374. |
|
|
Gower JC (1971). A general coefficient of similarity and some of its properties. Biometrics 27:857-874. |
|
|
IPGRI (1997) Descriptors for Yam (Dioscorea spp.). International Institute of Tropical Agriculture, Ibadan, Nigeria/International plant genetic resources institute, Available from View. |
|
|
Jaccard P (1908) Nouvelles recherches sur la distribution florale. Bull. Soc. Vaud. Sci. Nat. 44:223-270. |
|
|
Jamago JM (2003). Morpho-agronomic and molecular diversity of the Philippine mungbean (Vigna radiata L.) germplasm. Philipp J. Crop Sci. P. 174. |
|
|
Krzanowski WJ (1983). Distance between populations using mixed continuous and categorical variables. Biometrika 70:235-243. |
|
|
Lebot V (2009). Tropical root and tuber crops: cassava, sweet potato, yams and aroids. Wallingford: CABI. Crop Prod. Sci. Hortic. Series 17:413. |
|
|
Loko YL, Bhattacharjee R, Agre AP, Dossou-Aminon I, Orobiyi A, Djedatin GL, Dansi A (2016). Genetic diversity and relationship of Guinea yam (Dioscorea cayenensis Lam.–D. rotundata Poir. complex) germplasm in Benin (West Africa) using microsatellite markers. Genet. Resour. Crop pp. 1-15. |
|
|
Mahalanobis PC (1936). On the generalized distance in statistic. Proc. Nat. Inst. Sci. India 12:49-55. |
|
|
Matusita K (1955). Decision rules based on the distance, for problems of fit, two samples, and estimation. Ann. Math. Stat. 26:631-640. |
|
|
Mesquita AS (2001). Inhame na Bahia: a produção a caminho da competitividade. Bahia Agríc. 4(2):39-48. |
|
|
Moreira RFC, Silva SA, Silva AN, Da Silva MS, Cerqueira LS, Sousa CS, Sampaio Filho OM (2007). Descritores práticos para caracterização botânica de genótipos de inhame no Recôncavo baiano. In: Simpósio Baiano de Educação Ambiental, I . Cruz das Almas. Annals... Annals: SBEA. |
|
|
Nascimento WF, Siqueira MVBM, Ferreira AB, Ming LC, Peroni N, Veasey EA (2015). Distribution, management and diversity of the endangered Ameridian yam (Dioscorea trifida L.). Braz. J. Biol. 75:104-13. |
|
|
Nei M, Li WH (1979). Mathematical model for studying genetic variation in terms of restriction endonucleases. Proc. Natl. Acad. Sci. 76:5269-5273. |
|
|
Obidiegwu J, Loureiro J, Ene-Obong E, Rodriguez E, Kolesnikova-Allen M, Santos C, Muoneke C, Asiedu R (2009). Ploidy level studies on the Dioscorea cayenensis/ Dioscorea rotundata complex core. Euphytica 169:319-326. |
|
|
Padilla G, Cartea ME, Rodrigues VM, Ordás A (2005). Genetic diversity in a germoplasm collection of Brassica rapa Subs. Rapa L. from northwestern Spain. Euphytica 145:171-180. |
|
|
Pedralli G (2002). Distribuição geográfica e taxonomia das famílias Araceae e Dioscoreaceae no Brasil. In: Carmo CAS. Inhame e Taro: Sistemas de produção familiar. Vitória, ES: INCAPER. 289p. |
|
|
Pereira FHF, Puiatti M, Miranda GV, Silva DJH, Finger FL (2003). Caracterização agronômica da produção de rizomas de clones de taro. Hortic. Bras. 21(1):99-105. |
|
|
Pestana RKN, Amorim EP, Ferreira CF, Amorim VBO, Oliveira LS, Ledo CAS, Silva SO (2011). Agronomic and molecular characterization of gamma ray induced banana (Musa sp.) mutants using a multivariate statistical algorithm. Euphytica 178:151-158. |
|
|
Renyi A (1961). On measures of entropy and information. In Proceedings of the fourth Berkeley symposium on mathematical statistics and probability pp. 547-561. |
|
|
Santos ES (1996). Inhame (Dioscorea spp): aspectos básicos da cultura. João Pessoa: EMEPA-PB, SEBRAE. 158p. |
|
|
Santos ES, Cazé Filho J, Lacerda JT, Carvalho RA (2007a). Inhame (Dioscorea sp.) tecnologias de produção e preservação ambiental. Rev. Tecnol. Ciênc. Agropecu. 1(1):31-36. |
|
|
Santos ES, Fontinélli ISC, Lacerda JT, Matias EC, Barbosa MM (2007b). Sistema alternativo de produção de sementesde inhame (Dioscorea sp.). Rev. Tecnol. Ciênc. Agropecu. 1(2):19-24. |
|
|
Santos ES, Macêdo LS (2002). Tendências e pespectivas da cultura do inhame (Dioscorea sp.) no Nordeste do Brasil. In: Simpósio nacional sobre as culturas do inhame e do taro, 2, João Pessoa. Annals..., João Pessoa: SNIT. pp. 21-31. |
|
|
Santos ES, Macêdo LS, Matias EC, Melo AS (1998). Contribuição tecnológica para a cultura do inhame no Estado da Paraíba. EMEPAPB/MMA-PRONAF (EMEPA-PB. Documentos, 23). 84p. |
|
|
SAS Institute (2011). Statistical Analysis System. SAS Version 9.3.1 SAS Institute, Cary, North Carolina. |
|
|
Sheikh N, Kumar Y (2017). Morphological Characterization of Meghalayan Dioscorea spp. (yam), North East India. J. Agric. Sci. Technol.19(2):487-497. |
|
|
Silva LRG, Bajay MM, Monteiro M, Mezette TF, Nascimento WF, Zucchi MI, Pinheir JB, Veasey EA (2014). Isolation and characterization of microsatellites for the yam Dioscorea cayenensis (Dioscoreaceae) and cross-amplification in D. rotundata. Genet. Mol. Res. 13:2766-2771. |
|
|
Silva LRG, Mezette TF, Nascimento WF, Silva EF, Veasey EA (2016). Spatially structured morphological and molecular diversity among Dioscorea cayenensis and D. rotundata yam accessions. Plant Genet. Resour. https://doi.org/10.1017/S1479262115000647. |
|
|
Siqueira MVBM (2011). Yam: a neglected and underutilized crop in Brazil. Hortic. Bras. 29:16-20. |
|
|
Sudré CP, Gonçalves LSA, Rodrigues R, Amaral Júnior AT, Riva-Souza EM, Bento CS (2010) Genetic variability in domesticated Capsicum spp as assessed by morphological and agronomic data in mixed statistical analysis. Genet. Mol. Res 9:283-294. |
|
|
Vieira EA, Fialho JF, Silva MS, Faleiro FG (2007). Variabilidade genética do banco ativo de germoplasma de mandioca do cerrado acessada por meio de descritores morfológicos (Boletim de Pesquisa e Desenvolvimento, 129). Planaltina: Embrapa Cerrados 15 p. |
|
|
Ward Junior JH (1963). Hierarchical grouping to optimize an objective function. J. Am. Stat. Assoc. 58:236-244. |
|
Copyright © 2024 Author(s) retain the copyright of this article.
This article is published under the terms of the Creative Commons Attribution License 4.0