ABSTRACT
Lactic acid bacteria (LAB) are Gram-positive, non-spore-forming, catalase-negative cocci or rod-shaped bacteria that produce lactic acid as a major fermentation product. They are also involved in the production of fermented foods. They have applications in industry and human health, such as food preservation and probiotics. The aim of this research was to isolate, characterize, and classify indigenous lactic acid bacteria from fermented vegetable amaranth, a leafy vegetable native to Africa. The isolates' 16S rRNA gene was amplified using bacterial universal primers 27F and 1492R. From fermented vegetable amaranth, a total of 15 LAB were isolated were grouped into the genera Lactobacillus, Lactococcus, and Weissella based on 16S rRNA gene analyses. Lactobacillus plantarum dominated vegetable amaranth fermentation, accounting for 60% of all isolates.
Key words: Lactic acid bacteria, vegetable amaranth, fermentation, 16S rRNA.
Fermentation is one of the oldest food processing techniques. It is a technology that millions of people in the developing world depend on to preserve their food at prices that are affordable to the average consumer (Kalui et al., 2010). Fermented food items are a subset of foods that are distinguished by various carbohydrate breakdowns in the presence of probiotic microorganisms (Mulaw et al., 2019). Africa has a wide variety of fermented foods which include plant-based products from maize, sorghum, millet and cassava (Franz et al., 2014). It has been suggested that vegetables are a good source of lactic acid bacteria growth (Wu et al., 2020). Vegetables also have high contents of vitamins, minerals, dietary fiber, and antioxidant compounds (Wu et al., 2020).
Fermented foods are now known not only as a source of nutrients, but also as functional foods that offer health benefits against food-borne illnesses in addition to their nutritional value (Mulaw et al., 2019). Fermented vegetables contain a variety of microorganisms, including LAB, which produce lactic acid and, in rare cases, bacteriocins, which are essential for food preservation (Viridiana et al., 2018). In recent years, the probiotic potential of LAB isolated from fermented vegetable has been investigated (Viridiana et al., 2018). The microorganisms present during vegetable fermentation are very diverse and may considerably affect the quality and safety of the final product (Bautista-Gallego et al., 2020). The microbiota that is initially present in lactic fermentation processes comes primarily from the material, though other factors such as brines, ingredients used, and the industry's environment have an effect (Bautista-Gallego et al., 2020).
Lactic acid bacteria are a group of Gram-positive, aerotolerant, acid-tolerant, generally non-sporulating rod or cocci organisms that play an important role in food fermentation by inhibiting pathogenic microorganisms (Anjum et al., 2014). Lactic acid bacteria are present in a wide range of environments, including plant materials, various animal products, human and animal gastrointestinal and urogenital tracts, soil, and water (Ruiz Rodríguez et al., 2019). Lactic acid bacteria (LAB) use the Embden Meyerhof Parnas (EMP) pathway to ferment sugars and generate lactic acid as a final product (Anjum et al., 2014). Homofermentative lactic acid bacteria produce only lactic acid as a major product of fermentation, whereas heterofermentative lactic acid bacteria produce CO2, hydrogen peroxide, acetic acid, and alcohol in addition to lactic acid (Anjum et al., 2014). Organic acids and other low molecular weight substances produced by these microorganisms have been shown to improve food nutritional, sensory, technical, as well as protection and shelf-life properties (Oguntoyinbo et al., 2016a). Due to their versatile metabolism, LAB has been widely used as starter cultures and probiotics for these purposes (Naeem et al., 2012; Ruiz Rodríguez et al., 2019). Heterofermentative microorganisms usually perform the first steps of vegetable fermentation, producing lactic and acetic acids, which contribute significantly to the final product's flavor and aroma (Breidt et al., 2013). Then, because of their ability to produce lactic acid, which causes a greater decrease in pH but prevents the growth of other microbial classes, they are replaced by more acid-tolerant homofermentative microorganisms (Montet et al., 2014).
Amaranths are a diverse group of food crops with some grown as edible grains and others as edible leaves (Achigan-Dako et al., 2014). Amaranth belongs to the family Amaranthaceae (Zehring et al., 2015). Vegetable amaranth is one of the most widely consumed vegetables in Asia and Africa, and it plays a significant role in the supply of essential proteins and minerals (Achigan-Dako et al., 2014). The consumption of amaranth as a vegetable is mainly exclusive in Africa and Asia, whereas the grain is popular all over the world (Zehring et al., 2015). Amaranth leaves contain secondary plant metabolites such as saponins, flavonoids, betalains, tannins, etc. (Zehring et al., 2015).
Smallholder farmers in rural and peri-urban areas in Africa depend heavily on indigenous leafy vegetables for food protection. Post-harvest losses, on the other hand, may be as high as 50% due to poor production conditions (Abukutsa-Onyango, 2007). Although fermentation of vegetables, particularly cabbage, is common in Europe, fermentation of leafy indigenous vegetables is not yet common in Africa (Oguntoyinbo et al., 2016a). As a result, there appears to be a case for stepping up efforts in Africa to research and introduce this form of biological preservation system for leafy vegetables (Oguntoyinbo et al., 2016a). As a result, spontaneous fermentation was produced and monitored in this study to identify the LAB that enable fermentation and can be used as a viable approach for the preservation and enhancement of the protection of African leafy vegetables.
Sampling and preparation of sampling materials
Amaranthus dubius was obtained from Agrifood Organics, Ruiru, Kenya. The plant was cultivated for six to eight weeks (22-30°C). Hand-picking was used to harvest the vegetables, which were then shipped to the laboratory for processing. The leaves were destemmed, washed with tap water, and air-dried with paper towels on a clean and sterilized work bench.
Fermentation of vegetable amaranth
Fermentation was performed in 5-L stainless steel buckets. 1 kg of leaves and 3 L of salt and sugar solution were used. The solution consisted of a combination of salt and sugar, 3.0% each. Common table salt and retail sugar were purchased at local stores in Kenya. These components were sterilized by autoclaving for 15 min at 121°C. Weights were used to hold all plant material below the surface of the liquid. Inoculation and sampling were done in a sterile setting. The fermentations were done in duplicates at a constant temperature of 25°C (Stoll et al., 2021).
Sampling and analysis of fermentation brine
Samples were taken and analyzed for pH and microbial counts at 0, 24, 48, 72, and 144 h on MRS and M17 agar plates. The microbial diversity was investigated using amplicon sequencing of the 16S rRNA gene. 1 ml samples from different stages of vegetable amaranth fermentation brine were added to 9 ml quarter-strength Ringer's solution and vortexed to isolate LAB. To obtain the typical LAB associated with fermenting vegetable amaranth, the samples were further diluted in a 10-fold dilution sequence and 10 µl aliquots were spread-plated onto MRS agar (De Man, Rogosa, and Sharpe, M641) and M17 (M929) agar. Under aerobic conditions, plates were incubated at 30°C for 24-48 h (Stoll et al., 2021). For further characterization, colonies were selected at random from the highest dilution agar plates. The strains were then grown aerobically in MRS broth at 30°C and streaked to ensure purity. All media were purchased at Himedia (Mumbai, India). The isolates stock cultures were stored in MRS broth with 20% glycerol at -80°C (Abdou et al., 2018). The isolates were divided into groups based on phenotypic and biochemical characteristics, and their identity was confirmed through 16S rRNA gene sequencing.
Phenotypic characterization
Presumptive lactic acid bacteria were phenotypically identified using phase-contrast microscopy at 100x magnification (Shimadzu CX41, Japan), as well as standardized tests including catalase activity, gas output from glucose in MRS broth, growth at different NaCl concentrations (4 and 6.5%), and growth at different temperatures (15 and 45°C).
Determination of cell morphology and Gram status
Overnight cultures were introduced on microscopic slides and visualised under a light microscope at 100x magnification. Gram status was determined using 3% KOH as described by Mulaw et al. (2019).
Catalase test
Overnight cultures were introduced on a microscopic glass slide and two drops of 3% hydrogen peroxide added and thoroughly blend as described by Mulaw et al. (2019). The development of gas bubbles during a positive catalase test indicates that the test bacterium is producing catalase enzyme. The absence of gas bubbles indicates a negative catalase test.
Gas production from glucose fermentation
The aim of this test was to determine the homofermentative and heterofermentative properties of LAB isolates. Inverted Durham tubes with 1% glucose were used to measure CO2 output from glucose in modified MRS broth. Separately, 50 µl LAB culture was inoculated with 9 ml MRS broth in separate tubes containing 1% glucose and inverted Durham tubes. The test tubes were then incubated for 5 days at 30°C. The presence of gas bubbles in Durham tubes over the course of 5 days indicated that the isolates produced CO2 from glucose fermentation (Mulaw et al., 2019).
Growth at different temperatures
Growth at 15 and 45°C are the most frequently used for the classification of bacilli. To determine the growth at given temperatures, the MRS broth was used. 50 μl of overnight cultures were inoculated into 9-ml test media, incubated at 15 and 45°C and observed for five days for color and growth. Negative control was set using a 9-ml test tube containing the broth without inoculating with the LAB cultures (Mulaw et al., 2019).
Growth at different NaCl concentrations
The resistance of LAB isolates to various NaCl concentrations was tested. For this purpose, 4 and 6.5 % NaCl concentrations were used for testing. Test tubes containing 5 ml of modified MRS broth containing bromocresol purple indicator were prepared according to the concentrations needed and were inoculated separately with 50 μl of each overnight culture. The Test tubes were then incubated at 30°C for 5 days (Mulaw et al., 2019).
Genotypic characterization
Genomic DNA was extracted from overnight cell cultures grown in MRS broth using Quick-DNATM Fungal/Bacterial Miniprep Kit (Zymo Research, USA) according to the manufacturer’s protocols. The final DNA concentration and purification were determined using a NanoDrop spectrophotometer (PCRmax Lambda, Staffordshire, United Kingdom) and DNA quality was checked by 1% agarose gel electrophoresis. PCR amplification of 16S rRNA gene for presumptive LAB strains was done using bacterial universal primers 27 F:5`-AGA GTT TGA TCC TGG CTC AG-3` and 1492 R:5` GGT TAC CTT GTT ACG ACT T-3`. PCR was performed in a 50-µl reaction containing 25 µl One Taq® 2X Master Mix with standard buffer (New England Biolabs), 1 µl forward primer, 1 µl reverse primer, and 22 µl RNase free water. Then 49 µl of the mixture was added into a sterile PCR tube, and 1 µl of gDNA was added and used as a template. The condition of amplified gene fragment: pre-denaturation of the target DNA at 96°C for 4 min followed by 30 cycles at 94°C for 1 min, primer annealing at 51.5°C for 1 min, and 30 s and primer extension at 68°C for 8 min. PCR was completed with 10 min elongation at 68°C followed by cooling to 4°C. The reactions were carried out in a thermal cycler (ProFlex PCR systems). The size of the 16S rRNA gene PCR products was confirmed by electrophoresis on a 1% (w/v) agarose gel stained with GelRed and visualized using a Uvitec Cambridge gel documentation system (Uvitec, UK). PCR products were purified using the QIAquick PCR purification Kit (Qiagen, Germany) according to the manufacturer’s instructions. The purified amplicons were Sanger sequenced at Human Genomics Macrogen Europe (Macrogen Europe B.V, Amsterdam, Netherlands).
Phylogenetic analysis
The 16S rRNA gene sequences of the bacterial isolates were viewed for quality checks and edited using ChromasPro 2.1.8 software package. They were then compared with available standard sequences of bacteria lineages in the public nucleotide sequence databases in the National Center for Biotechnology Information (NCBI) using nucleotide blast (https://blast.ncbi.nlm.nih.gov/Blast.cgi) to find closely related bacterial 16S rRNA gene sequences. The 16S rRNA gene sequences of the isolates and those of the unknown closely related bacteria strains were aligned using MEGA (Molecular Evolutionary Genetics analysis) 7.0 software package (https://www.megasoftware.net) and phylogenetic trees were constructed using Maximum Likelihood method based on the Kimura 2-parameter model (Kimura M 1990) with MEGA (Molecular Evolutionary Genetics analysis) 7.0 software package (Kumar et al., 2016) (https://www.megasoftware.net). The tree topologies were evaluated using the bootstrap resampling method (Felsenstein, 1985) based on 1000 replicates. Escherichia coli was used as an outgroup.
Statistical microbial analysis
The microbial enumeration results were expressed as the mean and standard deviation of duplicate experiments of the counts and sampling sites using two-way ANOVA (GraphPad Prism version 8.4.2 software, GraphPad LLC, San Diego, California, USA), at a significance level of p<0.05.
Selection and enumeration of presumptive lactic acid bacteria
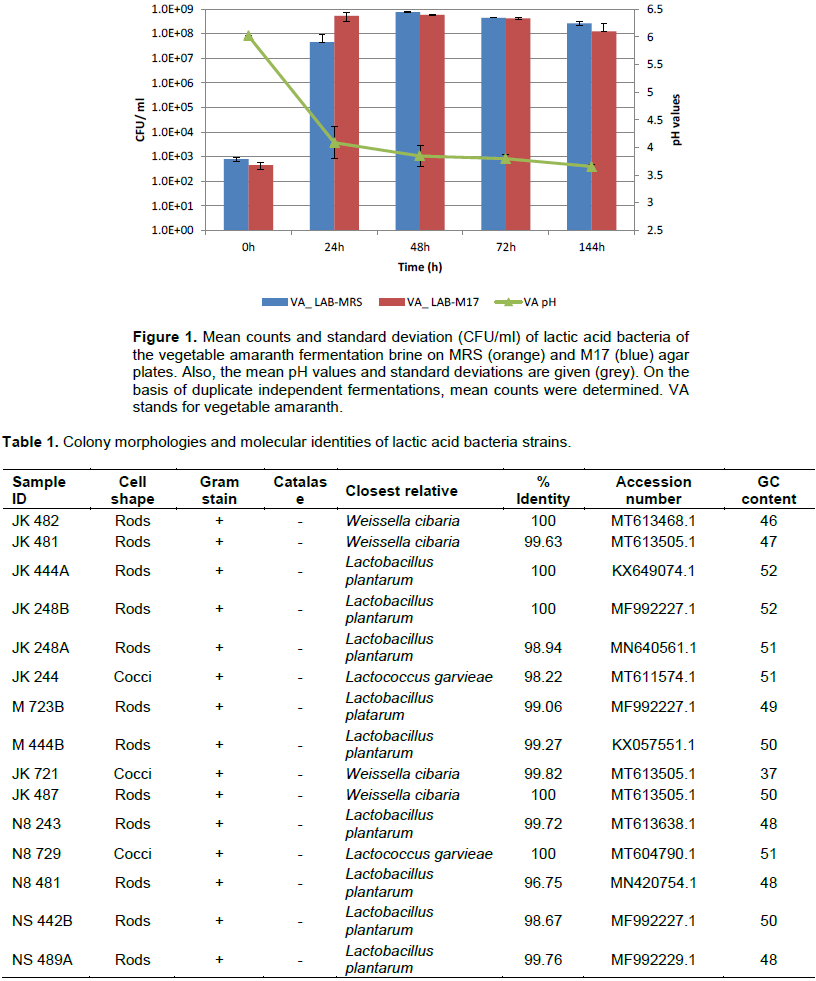
The microbial counts varied between the sampling points (Figure 1). The highest microbial counts were recorded between 24-48h. The results also showed a decrease in microbial counts between 72-144h. The ability of LAB to acidify during fermentation is reflected in the pH production. Within 24h of fermentation, the pH had reduced significantly from a pH 6.0 to pH 4.0. Within 48 h, the pH had reduced to pH 3.8, and by the end of the fermentation, it had dropped to pH 3.6. The counts of LAB on MRS and M17 agar were log 2 CFU/ml at the beginning of the fermentation and increased to log 7 and 8 CFU/ml after 24 h of fermentation, respectively. From 24 h to the end of the fermentation, the LAB counts ranged from log 7 to log 8.9 CFU/ml, suggesting that LAB constituted the majority of the isolates on these media (Figure 1). Classic macroscopic techniques of color, type, shape, and elevation of pure colonies were used to morphologically characterize presumptive LAB. Most colonies were able to grow within 2-4 days of incubation at 30°C. Gram's reaction, form, and catalase activity of bacterial species were also investigated. All of the isolates were Gram-positive and catalase-negative. The presumptive LAB strains are shown in Table 1.
Phenotypic characterization
In total, 15 Gram-positive, catalase-negative presumptive lactic acid bacteria were isolated from fermentation brine samples using MRS and M17 agar. Eight coccus-shaped isolates and seven rod-shaped isolates were among the presumptive lactic acid bacteria.
Morphological and physiological characterization
All isolates were identified according to their morphological and physiological properties (Table 1) and were able to grow at 4% NaCl salt concentration. Only 10 were found growing at 6.5% NaCl concentration. Examining the potential of the isolates to grow at 15 and 45°C showed that all isolates were able to grow at 15°C and only 12 isolates grew at 45°C. From the 15 isolates, 8 isolates produced gas from glucose. Thus, 8 isolates were found to be heterofermentative and 7 isolates were homofermentative (Kostinek et al., 2008).
Identification and phylogenetic analysis of isolates
Isolates were blasted on the Blastn search and showed similarities with percentage identities of 98 to 100% (Figure 2). Based on the Blastn search performed against the GenBank and phylogenetic analysis, nine isolates NS489A (MF992229.1), NS442B (MF992227.1), N8481(MN420754.1), N8243 (MT613638.1), M444B (KX057551.1), M723B (MF992227.1), JK248A (MN640561.1), JK 248B (MF992227.1) and JK444A (KX649074.1) were affiliated with Lactobacillus plantarum, Four isolates, JK482 (MT613468.1), JK481 (MT613505.1), JK721 (MT613505.1) and JK487 (MT613505.1) were affiliated with Weisella cibaria, and two isolates, JK244 (MT611574.1) and N8729 (MT604790.1) were affiliated with Lactococcus garvieae.
Fermented foods have a long history in Africa and are found all over the continent. Fermentation is a popular processing method for extending shelf life, increasing micronutrient availability, improving palatability, and improving digestibility (Wafula et al., 2016).
Amaranths, or Amaranthus spp., are a non-grass plant belonging to the Amaranthaceae family. Plants in this genus are important not only as vegetable and grain crops, but also as a source of vegetable protein for dry-land agriculture (Niveyro et al., 2012). Amaranth vegetable is one of the most popular leafy vegetable in Africa (Achigan-Dako et al., 2014). In many temperate and tropical regions, amaranth is widely grown as a green, leafy vegetable and grain crop. Amaranths have C4 photosynthesis and develop quickly in hot, dry environments. They also tolerate a wide range of unfavorable abiotic conditions, such as high salinity, acidity, or alkalinity, making them ideal for subsistence farming. Amaranth has the potential to have a positive effect on malnutrition if it is implemented (Maughan et al., 2011). Amaranths contain high contents of essential micronutrients such as calcium, iron, vitamin c, folic acid and b-carotene hence they have excellent nutritional value (Achigan-Dako et al., 2014).
Lactic acid bacteria are a group of microorganisms that produce lactic acid as a result of carbohydrate fermentation. They are normally considered micro-organisms with no pathogenic activities. They are widely used in the production of fermented dairy and non-dairy food products such as yoghurt (Streptococcus spp and Lactobacillus spp), cheese (Lactobacillus spp and Lactococcus spp), and sauerkraut (Leuconostoc spp) (Biosci et al., 2014). The vegetable amaranth was chosen for this study because it is an indigenous vegetable that is high in micronutrients, as well as being readily available and inexpensive. The minerals and vitamins content in AILVs are higher than those present in most exotic vegetables (Gido et al., 2017). In this study, we investigated the natural fermentation of the AILV vegetable amaranth in Kenya.
The study design was chosen to determine the dominance of lactic acid bacteria involved in the natural fermentation of this vegetable. Vegetable amaranth leaves fermented successfully in a fermentation brine containing 3% sugar and 3% salt. The aim of this study was to discover the diversity of the LAB population found in fermented vegetable amaranth. In present study, LAB counts were log 2 at the beginning of the fermentation and increased to log 7 and 8 respectively by 24h of fermentation. This was because, within the first 24h of the fermentation, the bacteria were at the exponential or log phase of growth whereby the cells are dividing and doubling in number. The LAB counts ranged from log 7 to log 8.9 from 24h of fermentation till the end of the fermentation. In line with this growth, there was a reduction in pH from pH 6.0 to pH 4.0 within the first 24h, and the pH further decreased to pH 3.6 by 144h of the fermentation. Lactic acid bacteria are acid-tolerant and produce acid and bacteriocin which reduces the pH and also inhibits the growth of pathogenic organisms (Masud and Anwaar, 2002; Oguntoyinbo et al., 2016a; Oluwajoba et al., 2013).A pH value of 4.2 or less is regarded as an important factor for food safety (Holzapfel, 2002).
Reduction of pH due to lactic acid bacteria fermentation between underneath 4.0 is critical to hinder the development of various pathogenic organisms (Oguntoyinbo et al., 2016a).
The 16S rRNA gene sequencing showed that the fermentation batches were characterized by a diverse microbiota consisting of strains belonging to the genus Lactobacillus, Weissella and Lactococcus (Oguntoyinbo et al., 2016a)and also found that spontaneous fermentation of African kale leaves was rather variable. The sequence data based on a constructed phylogenetic tree identified the isolates as L. plantarum, L. garvieae, and W. cibaria. Nine isolates were affiliated with L. plantaraum. Four isolates were affiliated with W. cibaria which have been described by Kang et al. (2016)as a Gram-positive, non-pore-forming, non-motile, hetero lactic acid-fermenting, and catalase-negative bacillus that cannot produce dextran from sucrose. Two isolates were affiliated with the genus Lactococcus with all two isolates associated with L. garvieae. Some genera of LAB isolated from this work like Lactobacillus and Weissella were also isolated in the previous study of Park et al. (2010)involving the use of 16S rRNA gene sequencing analysis to identify LAB diversity from fermented kimchi (a vegetable dish in Korea). According to the work carried out by other authors, Corsetti et al. (2001), Emerenini (2013), Sánchez et al. (2000), L. plantarum in plant materials was dominating the LAB flora. Similar to most other studies on non-dairy fermentation, L. plantarum strains were the LAB most dominant in our study (60% isolates). In addition, the study of LAB throughout the fermentation period clearly showed that L. plantarum-group strains occurred throughout the fermentation period (Huch et al., 2008). L. plantarum is the most common species found in fermented vegetables, owing to its ability to withstand the high saline and acidity content of fermented vegetables such as cucumber, sauerkraut, and olives (Behera et al., 2018). The results of the present study showed strains of Lactobacillus species from traditional fermented vegetable amaranth is important and more recurrent in vegetable amaranth fermentation and these findings can help us to have a better fermentation of this vegetable.
This work provides a microbiological and molecular study of fermented vegetable amaranth. The results of this study showed which strain of lactic acid bacteria is present in the fermentation of vegetable amaranth. Lactic acid bacteria were isolated from the fermentation of vegetable amaranth. The molecular results identified the lactic acid bacteria as L. plantarum, W. cibaria, and L. garvieae with L. plantarum being the most abundant species present. The purpose of this study was to use 16S rRNA sequencing to profile and taxonomically identify bacteria isolated from fermented vegetable amaranth. L. plantarum was the most dominant preceding lactic acid bacteria followed by W. cibaria and finally L. garvieae. According to the findings of this study, fermented vegetable amaranth contains a variety of lactic acid bacteria organisms. These bacterial strains could be used in a variety of industrial and commercial settings. As a result, further research is required to evaluate the isolates' functional properties as potential probiotics and starter cultures. Specifically, the molecular and metagenomic procedures would allow for a thorough examination of the function of LAB during fermentation, as well as microbial variations during processing, geographical and varietal effects.
This study did not use human or ethical subjects, and as such, ethical approval was not required.
The authors have not declared any conflicts of interests.
The authors appreciate the Pan African University Institute for Basic Sciences Technology and Innovation (PAUSTI) and Department of Food Science and Technology, Jomo Kenyatta University of Agriculture and Technology for provision of all laboratory facilities and assistance through the duration of this project. Authors’ are also grateful to the African Union for its financial support throughout the completion of this project.
REFERENCES
|
Abdou AM, Hedia RH, Omara ST, Mahmoud MAEF, Kandil MM, Bakry MA (2018). Interspecies comparison of probiotics isolated from different animals. Veterinary World 11(2):227-230.
Crossref
|
|
|
|
Abukutsa-Onyango M (2007). Seed production and support systems for African leafy vegetables in three communities in western Kenya. African Journal of Food, Agriculture, Nutrition and Development 7(3):1-16.
|
|
|
|
|
Achigan-Dako EG, Sogbohossou OED, Maundu P (2014). Current knowledge on Amaranthus spp.: Research avenues for improved nutritional value and yield in leafy amaranths in sub-Saharan Africa. Euphytica 197(3):303-317.
Crossref
|
|
|
|
|
Anjum N, Maqsood S, Masud T, Ahmad A, Sohail A, Momin A (2014). Lactobacillus acidophilus: Characterization of the Species and Application in Food Production. Critical Reviews in Food Science and Nutrition 54(9):1241-1251.
Crossref
|
|
|
|
|
Bautista-Gallego J, Medina E, Sánchez B, Benítez-Cabello A, Arroyo-López FN (2020). Role of lactic acid bacteria in fermented vegetables. Grasas y Aceites 71(2):1-9.
Crossref
|
|
|
|
|
Behera SS, Ray CR, Zdolec N (2018). Lactobacillus plantarum with functional properties: An Approach to increase safety and shelf-life of fermented foods. Hindawi BioMed Research International pp. 1-18.
Crossref
|
|
|
|
|
Biosci IJ, Ghafari E, Doosti A, Mokhtari-farsani A, Ghasemi P (2014). Genotypic and phenotypic study of Lactobacillus species isolated from traditional yogurt and cheese in southwest Iran. International Journal of Biosciences 5(10):155-162.
Crossref
|
|
|
|
|
Breidt F, Medina E, Wafa D, Pérez-Díaz I, Franco W, Huang HY, Kim, JH (2013). Characterization of cucumber fermentation spoilage bacteria by enrichment culture and 16S rDNA cloning. Journal of Food Science 78(3):M470-M476.
Crossref
|
|
|
|
|
Corsetti A, Lavermicocca P, Morea M, Baruzzi F, Tosti N, Gobbetti M (2001). Phenotypic and molecular identification and clustering of lactic acid bacteria and yeasts from wheat (species Triticum durum and Triticum aestivum) sourdoughs of Southern Italy. International Journal of Food Microbiology 64(1-2):95-104.
Crossref
|
|
|
|
|
Emerenini E (2013). Isolation and molecular characterization of lactic acid bacteria isolated from fresh fruits and vegetables using nested PCR analysis. British Microbiology Research Journal 3(3):368-377.
Crossref
|
|
|
|
|
Franz CMAP, Huch M, Mathara JM, Abriouel H, Benomar N, Reid G, Holzapfel W (2014). African fermented foods and probiotics. International Journal of Food Microbiology 190:84-96.
Crossref
|
|
|
|
|
Gido EO, Ayuya OI, Owuor G, Bokelmann W (2017). Consumption intensity of leafy African indigenous vegetables: towards enhancing nutritional security in rural and urban dwellers in Kenya. Agricultural and Food Economics 5(1).
Crossref
|
|
|
|
|
Holzapfel WH (2002). Appropriate starter culture technologies for small-scale fermentation in developing countries. International Journal of Food Microbiology 75:197-212.
Crossref
|
|
|
|
|
Huch M, Hanak A, Specht I, Dortu CM, Thonart P, Mbugua S, Franz CMAP (2008). Use of Lactobacillus strains to start cassava fermentations for Gari production. International Journal of Food Microbiology 128(2):258-267.
Crossref
|
|
|
|
|
Kalui CM, Mathara JM, Kutima PM (2010). Probiotic potential of spontaneously fermented cereal based foods - A review. African Journal of Biotechnology 9(17):2490-2498.
|
|
|
|
|
Kang BK, Cho MS, Park DS (2016). Red pepper powder is a crucial factor that influences the ontogeny of Weissella cibaria during kimchi fermentation. Scientific Reports 6:1-8.
Crossref
|
|
|
|
|
Kostinek M, Ban-Koffi L, Ottah-Atikpo M, Teniola D, Schillinger U, Holzapfel WH, Franz CMAP (2008). Diversity of predominant lactic acid bacteria associated with cocoa fermentation in Nigeria. Current Microbiology 56(4):306-314.
Crossref
|
|
|
|
|
Kumar S, Stecher G, Tamura K (2016). MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Molecular Biology and Evolution 33(7):1870-1874.
Crossref
|
|
|
|
|
Masud T, Anwaar K (2002). Role of lactic acid bacteria (LAB) in food preservation and human health - A review. Pakistan Journal of Nutrition 1(1):20-24.
Crossref
|
|
|
|
|
Maughan PJ, Smith SM, Fairbanks DJ, Jellen EN (2011). Development, characterization, and linkage mapping of single nucleotide polymorphisms in the grain amaranths (Amaranthus sp.). The Plant Genome 4(1).
Crossref
|
|
|
|
|
Montet D, Ray RC, Zakhia-Rozis N (2014). Lactic acid fermentation of vegetables and fruits. Microorganisms and Fermentation of Traditional Foods pp. 108-140.
|
|
|
|
|
Mulaw G, Sisay Tessema T, Muleta D, Tesfaye A (2019). In vitro evaluation of probiotic properties of lactic acid bacteria isolated from some traditionally fermented ethiopian food products. International Journal of Microbiology.
Crossref
|
|
|
|
|
Naeem M, Ilyas M, Haider S, Baig S, Saleem M (2012). Isolation characterization and identification of lactic acid bacteria from fruit juices and their efficacy against antibiotics. Pakistan Journal of Botany 44(SPL.ISS.1):323-328.
|
|
|
|
|
Niveyro SL, Mortensen AG, Fomsgaard IS, Salvo A (2012). Differences among five amaranth varieties (Amaranthus spp.) regarding secondary metabolites and foliar herbivory by chewing insects in the field. Arthropod-Plant Interactions 7(2):235-245.
Crossref
|
|
|
|
|
Oguntoyinbo FA, Cho GS, Trierweiler B, Kabisch J, Rösch N, Neve H, Franz CMAP (2016a). Fermentation of African kale (Brassica carinata) using L. plantarum BFE 5092 and L. fermentum BFE 6620 starter strains. International Journal of Food Microbiology 238:103-112.
Crossref
|
|
|
|
|
Oluwajoba SO, Akinyosoye FA, Oyetayo VO (2013). In Vitro Screening and Selection of Probiotic Lactic Acid Bacteria Isolated from Spontaneously Fermenting Kunu-Zaki. Advances in Microbiology 3(4):309-316.
Crossref
|
|
|
|
|
Park JM, Shin JH, Lee DW, Song JC, Suh HJ, Chang UJ, Kim JM (2010). Identification of the lactic acid bacteria in kimchi according to initial and over-ripened fermentation using PCR and 16S rRNA gene sequence analysis. Food Science and Biotechnology 19(2):541-546.
Crossref
|
|
|
|
|
Ruiz Rodríguez LG, Mohamed F, Bleckwedel J, Medina R, De Vuyst L, Hebert EM, Mozzi F (2019). Diversity and functional properties of lactic acid bacteria isolated from wild fruits and flowers present in northern Argentina. Frontiers in Microbiology.
Crossref
|
|
|
|
|
Sánchez I, Palop L, Ballesteros C (2000). Biochemical characterization of lactic acid bacteria isolated from spontaneous fermentation of "Almagro" eggplants. International Journal of Food Microbiology 59(1-2):9-17.
Crossref
|
|
|
|
|
Stoll DA, Wafula EN, Mathara JM, Trierweiler B, Kulling SE, Huch M (2021). Fermentation of African nightshade leaves with lactic acid bacterial starter cultures. International Journal of Food Microbiology 342:109056.
Crossref
|
|
|
|
|
Viridiana CR, Lidia DA, Audry PL, Humberto HS (2018). Lactic acid bacteria isolated from vegetable fermentations: probiotic characteristics.
Crossref
|
|
|
|
|
Wafula EN, Franz CMAP, Rohn S, Huch M, Mathara JM, Trierweiler B, Becker B (2016). Fermentation of African Leafy Vegetables to Lower Post-Harvest Losses , Maintain Quality and Increase Product Safety. African Journal of Horticultural Science 9:1-13.
|
|
|
|
|
Wu C, Li T, Qi J, Jiang T, Xu H, Lei H (2020). Effects of lactic acid fermentation-based biotransformation on phenolic profiles, antioxidant capacity and flavor volatiles of apple juice. LWT 122:109064.
Crossref
|
|
|
|
|
Zehring J, Reim V, Schröter D, Neugart S, Schreiner M, Rohn S, Maul R (2015). Identification of novel saponins in vegetable amaranth and characterization of their hemolytic activity. Food Research International 78:361-368.
Crossref
|
|