Full Length Research Paper
ABSTRACT
Yield losses in food crops due to plant pathogenic bacteria are significant and increasing over the years. The increasing losses caused by bacterial plant pathology are explained by the emerging resistance of bacteria to the chemical agents used in plant protection. Moreover, these chemical agents harm the environment through residue accumulation leading to soil pollution and the perturbation of the soil's inner ecosystem. The most important bacteria causing plant pathology belong to the genera of Pseudomonas, Ralstonia, Agrobacterium, Xanthomonas, Erwinia, Xylella, Pectobacterium, and Dickeya. However, in Côte d'Ivoire, only the Ralstonia species have been identified. Therefore, this study aims to identify plant pathogenic bacteria present in market garden plants in Côte d'Ivoire. Three sites in the cities of Anyama, Abidjan, and Bingerville were selected for the sampling and the detection of Pseudomonas syringea, Erwinia carotovora, Clavibacter michiganensis, Ralstonia solanacerum, and Xanthomonas campestris. The samples consisted of healthy and affected plant leaves and soils. In brief, 70 bacterial strains were isolated and phenotypically identified in this study. Among them, we noticed that 20% were isolated from the leaves and 80% from the soil. Regarding the bacterial species, C. michiganensis (37.14%), E. carotovora (18.57%), R. solanacerum (15.71%), X. campestris (14.28%), and P. syringae (11.42%) were identified. The molecular identification has confirmed the identification of the 5 plant pathogenic bacteria within all the studied sites. To the researchers’ knowledge, this study is the first to describe the identification of P. syringea, E. carotovora, C. michiganensis, and X. campestris isolated in plant crops in Côte d’Ivoire.
Key words: Phytobacteria, vegetable plants, PCR
INTRODUCTION
Vegetables contribute to more than 33% of world agricultural production and employ 800 million people (Kanda et al., 2014). Tomato (Solanum lycopersicum), red pepper (Capsicum spp.), and, eggplant (Solanummelongena) are among the 40 most cultivated plant species in the world (FAO, 2008). In sub-Saharan Africa, market gardening represents an important sector of activity because of the nutritional material produced, but also because of the source of income that it provides (Djiéto-Lordon, 2007). In Côte d'Ivoire, the market gardening of vegetables represents an average of 27% of the gross national product (GDP). In addition, the demand for vegetables such as tomatoes, eggplants, and pepper is increasing due to population growth. Then, farmers are intensifying their production to cover the needs. However, this production faces many biotic constraints such as bacterial diseases causing high losses, especially in tropical climates (Lebeau, 2010). The plant pathogenic diseases led to the decrease in agricultural yield and the increase in the prices of the products on the market. Therefore, there is need to identify bacteria responsible for plant pathogenic diseases in Côte d’Ivoire to propose a solution to overcome the problem. According to their scientific and economic importance in the world, five bacterial species were highlighted (Mansfield et al., 2012). Firstly, Ralsotonia solanacearum is responsible for bacterial wilt in tomatoes, eggplants, and potatoes. The bacterium, initially, infects the roots and then, invades the vascular system of the plants (Nakahara et al., 2021). Next, Pseudomonas syringae an epiphytic bacterium that survives on weed roots, asymptomatic plants as well as seeds is known to cause symptoms of bacterial speckling (black dot surrounded by a yellow halo) (Canzoniere et al., 2021). Xanthomonas campestris, for its part, is responsible for black rot and causes vascular disease in some plants or leaf spots in others (Vicente and Holub, 2013). Following this, Clavibacter michiganensis spreads through plant vessels and causes symptoms such as wilting, stem canker, vascular discoloration, and cell habit (Yim et al., 2012). The contaminated seeds by C. michiganensis remain the main mean of bacterial propagation. Finally, Erwinia carotovora enters the plant through wounds. The plant walls are then degraded and the tissues are macerated by pectolytic enzymes causing soft rotting of stems and fruits (Boumaaza et al., 2018). Regarding the economical and nutritional implications of bacterial plant diseases, this study aimed to detect the presence of cited bacteria that can cause infections in tomato, eggplant, and chili pepper.
MATERIALS AND METHODS
Samples collection
The sampling sites chosen are fields where organic culture is practiced. Eggplant and pepper plant samples were collected from site 1, located in a forest of the city of Anyama and near a waterway. Tomato plant samples were collected from site 2, located in the city of Abidjan. Finally, eggplant and pepper plant samples were also collected on site 3 in the outskirt of Bingerville city (Figure 1). For each plant, samples of diseased and healthy leaves were collected and at the base of each plant, soil samples were also collected. All in all, for each site, 3 soil samples and 3 leaves samples were collected per plant. Soil samples (10 g) were taken at 15cm depth (Popoola et al., 2015). The leaves were cut off at the base of the petiole. Healthy and diseased leaves were collected at the rate of 10 leaves per plant (Gracein et al., 2012). Plant samples collected were carefully bagged, shipped to the laboratory, and processed immediately or stored at 4°C and processed within 48 h.

Pre-treatment
Small pieces of leaves were cut aseptically; the surface of the leaf was sterilized in 70% alcohol and washed in three series of sterile distilled water to remove traces of alcohol. The leaves were suspended in tubes containing 3 ml sterile distilled water for 15 to 20 min (Gracein et al., 2012; Nakahara et al., 2021). One gram of soil sample was weighed and added to a test tube containing 10 ml of physiological water (9% NaCl). Then, the tubes were shaken for 30 min, 50 rpm at room temperature to allow the separation of bacteria from the soil (Popoola et al., 2015 modified).
Isolation and characterization of phytobacteria
Isolation
A serial decimal dilution of the bacterial suspensions obtained after pretreatment was carried out in 9 ml of sterile distilled water when the water contained in the tubes became slightly cloudy. Then, 1 ml of the diluted bacterial cell suspension was poured onto sterilized Petri plates containing nutrient agar King B or YPGA. The inoculated plates were incubated at 28° for 24 to 48 h. Depending on the appearance, coloring, and morphology of the bacterial colony; an isolated colony was picked and plated again on nutrient agar using a Pasteur pipette (Table 1). This step was repeated three times to allow purification of the isolated strain (Chartier, 2005; Amkraz et al., 2010; Gracein et al., 2012; Huynh et al., 2019).

Bacterial detection by polymerase chain reaction
The DNA extraction of isolated bacteria was performed using Qiagen® DNA extraction kit following the manufacturer’s recommendations. Fives colonies of bacteria were diluted in 100µl water (Qiagen, 2003).
The PCR analysis was performed to amplify conserved genes for each phytobacterium. The reaction mix contained 1XFirepolMasterMix (SolisBiodyne), 16 µl molecular water, 5 µl of DNA extract, and 0.5 µM specific primers for each bacterium (Table 2). For amplification, initial denaturation was carried out at 95 °C for 10 min, followed by 32 cycles of denaturation at 95°C (30 s), annealing at 54°C (30 s), and elongation at 72°C (1 min). Finally, the elongation of 5 mn was carried out. The amplification program was conducted on Applied Biosystems (9700 PCR System thermocycler). PCR products were visualized using 1.5% (wt/vol) agarose gel electrophoresis and a 100-bp DNA ladder (Promega).

Bacterial activity tests
Bacterial growth in vitro
The bacterial count makes it possible to know the exponential growth time of the bacteria used. This count is done at different times (0, 2, 4, 6, 8, and 24h). A stock solution of fresh phytobacteria is serially diluted up to 10-8 and then plated on King B or LPGA agar. The tests were repeated in triplicate. The dishes were incubated at 28°C for 24 h.
Bacterial pathogenicity test in vivo
One-month-old tomato plants were inoculated with a fresh culture of the following bacteria: R. solanacerum, E. carotovora, X. campestris, C. michiganensis, and P. syringea. The bacterial strains used are those that have been confirmed by PCR. Inoculation was done by adding 500µl of fresh bacterial culture to the leaf surface of each plant. The inoculated plants are observed until the appearance of the first symptoms (Table 1, Figure 9). The inoculated plants were re-isolated on agar and compared to the base strains as follows. Leaves from each plant were cut (3 leaves per plant) and placed in tubes containing 2ml of sterile distilled water for 1 hour. Dilutions in physiological water were then made and 10µl spots were deposited on the agar incubated at 28°C for 24 h. All the tests were repeated three times for each bacterium (Kumar et al., 2017 modified).
RESULTS
Collection of samples and bacterial identification
A total of 70 bacteria were isolated. The strains isolated from site 1 are 23 in number, 25 from site 2, and 22 from site 3. Among them, 20% were isolated from the leaves and 80% from the soil. According to the bacterial species, there were 38.57% of Clavibacter sp; 17.14% Erwinia sp, 14.728% Ralstonia sp; 12.85% Xanthomonas sp, and 14.42% Pseudomonas sp. 36% of the bacterial strains were isolated from site 2, while for sites 1 and 3 we had 33 and 31% of isolated bacteria respectively (Table 3), (Figures 2 and 3). The five investigated bacteria were recovered in the 3 sites.

Quantification of genomic DNA
Genomic DNA was extracted from 70 strains. The amount of nucleic acid was quantified using Nanodrop oneC instruments. Indeed, the highest amounts of DNA were identified in X. campestris followed by strains 26 and 15 of C. michiganensis then strains 10 and 8 of Ralstonia. The highest concentrations are 218, 246, 172, 165.5 and 165.1 µg/ml. The purity of the samples was between 1.4 and 2.1. The table shows the concentration and purity of the PCR-positive strains (Table 4).

Molecular characterization
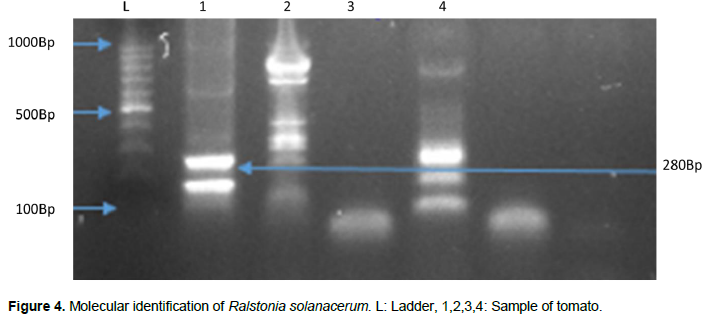
Genomic amplification by PCR made it possible to obtain 271 bp fragments for C. michiganensis representing the 16S-23S gene. Products of 280 bp for R. solanacearum and 435 bp for E. carotovora were both obtained targeting the 16S-23S gene. P. syringae targets the HrpZpst gene with a fragment size of 525 bp, while the X. campestris (hrp) gene has a fragment size of 304 bp. For molecular characterization by PCR, we identified 28.57% positive phytobacteria. According to the bacterial species, there is 35% of C. michiganensis, 5% E. carotovora, 20% R. solanacearum, X. campestris and P. syringae (Figures 4 to 9).


Bacterial growth curve
The number of colony-forming units (CFU) was evaluated at different dilutions of isolates at different times. The isolates used are the PCR positive isolates. The number of colony-forming units (CFU) was assessed at different dilutions of isolates at different time points. The isolates used are the positive PCR isolates. The highest bacterial growth was observed in E. carotovora; followed by one of the Ralstonia strains. Two of the strains of Clavibacter had similar bacterial growth. They could be the same strain or a neighboring strain. Clavibacter strain 18 has much weaker growth than the previous two strains. All like the two strains of Xanthomonas and Erwinia which could come from distant strains (Figure 10).

Pathogenicity test
Pathogenicity tests showed that the selected strains were able to induce infection in tomato plants. The first symptoms appeared 14 days after inoculation for R. solanacearum, E. carotovora, and X. campestris. Early symptoms of R solanacearum were characterized by yellowing and wilting of leaves. Erwinia induces yellowing of the leaves while small yellow to brown spots have been observed on the leaves of plants infected with X. campestris. C. michiganensis and Pseudomonas appear one month after inoculation in the plant. Symptoms of C. michiganensis were manifested by the appearance of a burning color on some leaves as well as their wilting. One appearance of leaves infected by Pseudomonas was characterized by the appearance of yellowish to brown spots on the leaves. The bacterial count of each strain was assessed weekly after inoculation. The bacteria concentrations obtained were listed in Figures 10 and 11. The figure showed that from 0 to 4 days, the bacterial concentration of all bacteria was almost zero. Between 14 and 10 days, the concentration of bacteria increased slightly. From day 10, bacterial growth of Clavibacter; Xanthomonas, and Erwinia on leaves increased exponentially. As for Ralstonia and Pseudomonas, growth remained low throughout the experiment.

DISCUSSION
This study confirms the presence of phytopathogenic bacteria in leaf and soil samples from the sites. A collection of 70 bacterial strains was isolated. The bacteria isolated from site 1 are 33%. The results corroborate those of Kumar et al, 2017 in their study which shows that the rhizospheric bacteria were a mixture of antagonists and neutral and mutualistic pathogens. Site 1's cultivation system was organic and chemical-free. The constituent elements of the rhizosphere have therefore been preserved. And there is a close connection between subsoil microbes and aboveground components of the plant ecosystem (Kumar et al., 2017). Previously used as a household waste dump 36% of the phytobacteria were isolated from site 2. Indeed, this waste was the growth biotope of several microorganisms of bacterial, viral, and parasitic origin. And Cissé's study demonstrated that a biotope where contamination is high presents a high risk for cultivated plants (Cissé, 1997). C. michiganensis responsible for bacterial canker, was the most isolated phytobacteria on the three sites with 38.57% confirmation. The results coincide with those of Ftayeh and Nandi who state that Clavibacter is a devastating emerging disease for crops. It is a serious disease in the world and new epidemics have recently been reported in several countries (Ftayeh et al., 2011) hence its strong presence in these soils has
caused significant crop losses. It is also considered one of the most devastating plant diseases of crops (Nandi et al., 2018). It is followed by E carotovora preferentially isolated on site 2 at 17.14%. The results are contrary to the results of a previous study by Benada which showed that Erwinia sp is a bacterium that preferentially infects fruits and potatoes (Benada et al., 2019). Damaged fruits and potatoes deposited on the plot of site 2 formerly used as a dump could be the cause of the appearance of this bacterium. And the insect vectors par excellence have allowed the spread of this bacterium in plants (MHMED, 2019).
One of the most important bacteria in peppers and crucifers is Xanthomonas campestris. It is seed-borne and can survive in weeds and plant debris for months. These matrices are a biotope for surviving Xanthomonas (Laala et al., 2021). Xanthomonas campestris was isolated with a percentage of 12.85% in tomatoes. A previous study isolated xanthomas in rice and it was Xanthomonas oryzae pv. Oryzicole in Ivory Coast (Diallo et al., 2010). However, strains of Xanthomonas campestris have just been isolated for the first time in Côte d'Ivoire. At the level of molecular characterization, 50% of Pseudomonas isolates were PCR positive. Although the hrp gene is unknown in virulence and pathogenicity, this region is essential for the production of symptoms in the host (Zaccardelli et al., 2005). It is considered a stable gene using 16S-
The isolates of C. michiganensis (21.42%) are Pcr positive. ITS region sought for molecular confirmation of Clavibacter is a relatively variable area. Yim et al. (2012) showed a difference of more than 50% between the ITS regions of tomato and pepper crops. And a diversity of genetic variation exists between Clavibacter populations in several countries (De Leon et al., 2009). Our pathogenicity test results showed that R. solanacerum; E. carotovora and X. campestris induce symptoms on tomato plants. The first symptoms appeared after 14 days. And according to the literature the first symptoms appear after an infection time of 14 days (Iiyama et al., 2021). These bacteria are preferentially present in the rhizosphere. They, therefore, infect the plant through the roots and invade the vascular system. The bacteria grow and produce extracellular polysaccharides which cause obstruction of the first symptoms on the plant and the death of the latter (Nakahara et al., 2021). The Ralstonia strains used induced infection in tomato plants. It could therefore be race 3. Indeed, according to a study by (Kumar et al., 2017), the strains capable of infecting Solanaceae are part of race 3.
After one month of infection, all plants infected with C. michiganensis and P. syringea showed symptoms. These results are consistent with those of Yim et al. (2012) which show 25 days after inoculation, the plants show symptoms and 80% of the plants die. This study confirmed the presence of phytobacteria in plants. 70 strains of phytopathogenic bacteria were isolated in three sites (rural, semi-rural, and urban sites). And 20 strains have been confirmed by molecular diagnosis. E. carotovora strains X. campestris C. michiganensis and P. syringea were isolated for the first time in this study. C. michiganensis is the most predominant strain in the tomato and eggplant plant. E. carotovora and R. solanacerum are abundant at all sites. These strains induced in vivo infection tests with major leaf symptoms. Isolated bacteria are responsible for various infections resulting in huge production losses. To compensate for these losses, the use of biopesticides such as agriphages is a godsend, especially since their isolation is inexpensive. The isolation of agriphages would then be a considerable alternative to reduce production losses due to phytopathogens.
CONFLICT OF INTERESTS
The authors have not declared any conflict of interests
REFERENCES
|
Amkraz N, Boudyach EH, Boubaker H, Bouizgarne B, Aoumar AA (2010). Screening for fluorescent pseudomonades, isolated from the rhizosphere of tomato, for antagonistic activity toward Clavibacter michiganensis subsp. michiganensis. World Journal Microbiology and Biotechnology 26(6):1059-1065. |
|
|
Boumaaza B, Boudalia S, Khaladi O, Guessas B, Bella A (2018). Variability of aggressiveness and virulence of Erwinia carotovora subsp. carotovorum causing the soft rot on potato tubers in the western of Algeria. International Journal of Plant Biology 9(1):7568. |
|
|
Canzoniere P, Francesconi S, Giovando S (2021). Antibacterial activity of tannins towards Pseudomonas syringae pv. tomato, and their potential as biostimulants on tomato plants. The international Journal of the Mediterranean Phytopathological Union 60:23-36. |
|
|
Cissé (1997). Impact sanitaire de l'utilisation d'eaux polluées en agriculture urbaine. Cas du maraîchage à Ouagadougou (Burkina Faso). Ecole polytechnique de losanne. 447p. |
|
|
Chartier R (2005). Méthodes d'appréciation du comportement variétal vis-à-vis des bioagresseurs. Cahier des. |
|
|
Djiéto-Lordon C, Aléné DC, Reboul JL (2007). Contribution à la connaissance des insectes associés aux cultures maraichères dans les environs de Yaoundé -Caméroun. Cameroun Journal Biological and Biochemical Sciences 15:1-13. |
|
|
Food and Agriculture Organization (FAO) (2008). La flambée des denrées alimentaires: faits, perspectives, effets et actions requises. Document HLC/08/INF/1 préparé pour la Conférence de haut niveau sur la sécurité alimentaire mondiale: Les défis du changement climatique et des bioénergies, 3-5 juin 2008, Rome. |
|
|
Ftayeh RM, Tiedemann AV, Rudolph KWE (2011). A New Selective Medium for Isolation of Clavibacter michiganensis subsp michiganensis from Tomato Plants and Seed A New Selective Medium for Isolation of Clavibacter michiganensis subsp. michiganensis from Tomato Plants and Seed. The American Phytopathological Society 101(11):1355-1364. |
|
|
Guessan CAN, Abo K, Fondio L, Chiroleu F, Lebeau A, Poussier S, Wicker E, Koné D (2012). So Near and Yet so Far?: The Specific Case of Ralstonia solanacearum Populations from Côte d' Ivoire in Africa. Phytopathology 102. |
|
|
Gracein DH, Herin D, De Britto AJ, Kumar R, Jeya PB (2012). Detection and identification of Xanthomons cempestris pv. centellae on leaves of Centella asiatica collected in Tamilnadu. Asian Journal of Pharmaceutical and Clinical Research 5(1):111-113. |
|
|
Huynh NT, LeUyen T, Trinh QP, Tran TT, Luu TD, Nguyen TTN (2019). Isolation and Virulent Evaluation of Ralstonia solanacearum cause the Bacterial Wilt in Chrysanthemum (Chrysanthemum Sp.) from Mekong Delta and Lam Dong Province. Biological Forum an international Journal 11(1):101-106. |
|
|
Iiyama K, Imamura M, Inoue T, Kyaw HWW, Yano K, Horita M, Tsuchiya K, Furuya N (2021). Pathogenicity evaluation of Ralstonia pseudosolanacearum race 4 on ginger by leaf-clipping inoculation. Journal of General Plant Pathology 87(5):269-272. |
|
|
Kanda M, Akpavi S, Wala K, Djaneye-Boundjou G, Akpagana K (2014). Diversité des espèces cultivées et contraintes à la production en agriculture maraichère au Togo. International Journal of Biological and Chemical Sciences 8(1):115-127. |
|
|
Kiran R, Kandan A, Kumar P, Singh D, Akhtar J, Singh B, Dubey SC (2019). Development of species-specific primers for detection of xanthomonas campestris pv. campestris causing black rot of crucifers. Journal of Environmental Biology 40(1):105-110. |
|
|
Kumar S, Hamsaveni N, Gowda PHR (2017). Isolation and Characterization of Ralstonia solanacearum Causing Bacterial Wilt of Solanaceae Crops. International Journal of Current Microbiology and Applied Sciences 6:1173-1190. |
|
|
Laala S, Cesbron S, Kerkoud M, Valentini F, Bouznad Z, Jacques MA (2021). Characterization of Xanthomonas campestris pv . campestris in Algeria. Phytopathologia Mediterranea 60(1):51-62. |
|
|
Lebeau A (2010). Resistance de la tomate, l'aubergine et le piment à Ralstonia solanacearum:interactions entre les géniteurs de résistance et la diversité bactérienne, caractérisation et cartographie des facteurs génétiques impliqués chez l'aubergine. |
|
|
Mansfield J, Genin S, Magori S, Citovsky V, Sriariyanum M, Ronald P, Dow M, Verdier V, Steven VB, Machado MA, Toth I, Salmond G, Foster GD (2012). Top 10 plant pathogenic bacteria in molecular plant pathology. Molecular Plant Pathology 13(6):614-629. |
|
|
Muhamed B (2019). Caractérisation phénotypique et génotypique d'Erwinia sp pathogène et essaie de lutte biologique Soutenue. https://theses.univ-oran1.dz/document/TH5096.pdf |
|
|
Nakahara H, Maehara A, Mori T, Matsuzoe N (2021). Selection of Eggplant Cultivars and Combination with Graft Cultivation for Effective Biological Control of Vascular Wilt Diseases Using a Phenotypic Conversion Mutant of Ralstonia solanacearum. The Japanese Society for Horticultural Science P 1. |
|
|
Nandi M, Macdonald J, Liu P, Weselowski B, Yuan ZC (2018). Clavibacter michiganensis ssp. michiganensis: bacterial canker of tomato, molecular interactions, and disease management. Molecular Plant Pathology 19(8):2036-2050. |
|
|
Photchanachai S, Cheevadhanarak S, Phokum C, Jitareerat P (2006). Detection and classification of soft rot Erwinia of vegetables in Thailand by DNA polymerase chain reaction. In IV International Conference on Managing Quality in Chains-The Integrated View on Fruits and Vegetables Quality 712:917-926. |
|
|
Popoola AR, Ganiyu SA, Enikuomehin OA, Bodunde JG, Adedibu OB, Durosomo HA, Karunwi OA (2015). Isolation and Characterization of Ralstonia solanacearum Causing Bacterial Wilt of Tomato in Nigeria. Nigerian Journal of Biotechnologie 29:1-10. |
|
|
Qiagen (2003). QIAamp ® DNA Mini Kit and QIAamp DNA Blood Mini Kit Handbook For DNA purification from Whole blood Buffy coat Body fluids Lymphocytes Cultured cells Tissue Swabs Dried blood spots February 2003. |
|
|
Vicente JG, Holub EB (2013). Pathogen profile Xanthomonas campestris pv. campestris (cause of black rot of crucifers) in the genomic era is still a worldwide threat to brassica crops. Molecular Plant Pathology 14(1):2-18. |
|
|
Yim K, Lee H, Kim J, Lee S, Cho J, Cha J (2012). Characterization of phenotypic variants of Clavibacter michiganensis subsp. michiganensis isolated from Capsicum annuum. European Journal of Plant Pathology 133(3):559-575. |
|
|
Zaccardelli M, Spasiano A, Bazzi C, Merighi M (2005). Identification and in planta detection of Pseudomonas syringae pv. tomato using PCR amplification of hrpZPst. European Journal of Plant Pathology 111(1):85-90. |
|
Copyright © 2024 Author(s) retain the copyright of this article.
This article is published under the terms of the Creative Commons Attribution License 4.0