Mastitis is one of the most important diseases in dairy milk systems. Molecular studies of the principal agents that are involved, as well as tests of bacterial sensitivity to plant extracts, are currently the most common forms of research related to this disease. The aim of the present study was to molecularly characterize isolates from cases of subclinical mastitis and to investigate the production of biofilm associated with extracts from Hymenaea martiana. The phytochemical screening of the crude ethanolic extract (CEE) confirmed the presence of phenolic substances, flavonoids, steroids and terpenoids. The antimicrobial susceptibility profile of the isolates that were assessed exhibited variations, with a notably high sensitivity to gentamicin. After the addition of the extract, 77.3, 81.8 and 86.3% of the isolates exhibited a reduction in biofilm production in the ethyl acetate, chloroform and hexane fractions, respectively. An extensive production of biofilm can be observed in the isolates that were not in contact with the natural extract. However, when in contact with the plant extract, there was a reduction in biofilm production, thereby demonstrating the therapeutic potential of the natural extract of H. martiana in mastitis caused by Staphylococcus spp.
Mastitis is one of the main diseases in dairy herds, considering the high costs resulting from falling milk production and quality, and also due to the costs that involve the control of the mastitis in dairy farms. This infirmity is characterized by an inflammatory process of the mammary gland, commonly caused by infectious microorganisms. Regarding the etiology in dairy goats, intramammary infections are mostly caused by several species of Coagulase-negative Staphylococcus (SCN) (McDougall et al., 2014; Gelasakis et al., 2016).
Among the several virulence factors, biofilm production is one of the main responsible factors for the persistence of intramammary infections. It is a structure formed by the organization of bacterial cells as a biopolymer matrix that surrounds the bacteria and attaches them to a surface. Bacterial adhesion to epithelial cells is usually the first stage of the infectious process and prevents the elimination of bacteria during milking. Biofilm formation occurs in successive stages, with its beginning characterized by the attachment of bacteria to a solid surface, where they multiply, and groups of cells accumulate on multilayers, resulting in the formation of a bacterial community. Biofilm protects the bacteria against the immune response of the host and the action of antimicrobial agents (Salaberry et al., 2015; Algharib et al., 2020).
Regarding the treatment of subclinical mastitis, besides the above-mentioned virulence factor, it is also important to take into account the cost, the drug elimination time, and the milk loss during the therapy period. The use of antimicrobials constitutes the main form of treatment for mastitis cases in the property. However, the high cost and the bacterial resistance to those compounds have led researchers to search for new alternatives to control this infirmity. The combination of natural compounds with commercial drugs has been a vast field for scientific research and may contribute to minimizing the resistance impact to pathogens (Mushtaq et al., 2018; Procópio et al., 2019).
Considering that conventional antimicrobial therapies present the disadvantage of selecting resistant bacteria, other mechanisms aiming at the control of infirmities have been sought. Therefore, controlling only the virulence of bacteria, but not their growth, seems to be a viable alternative. Plants might offer opportunities for the discovery of potentially bioactive molecules against the virulence factors of pathogenic organisms. Some studies have evaluated the ability of plant extracts in interfering with the mechanisms related to virulence factors, such as the potential of inhibiting or destructing biofilms or even interfering with quorum-sensing systems (QS) (Nazzaro et al., 2013; González-Ortiz et al., 2014).
According to Boniface et al. (2017), the plants of the genus Hymenaea are used in South America and Asia to treat several infirmities, among them, those caused by bacterial pathogens and parasites. The flavonoid astilbin has been reported as the main phenolic compound in this genus (Silva et al., 2012). Extracts containing high contents of phenolic compounds may be directly related to the antimicrobial activity (Dimech et al., 2013). In this perspective, this work aimed to perform the molecular characterization, evaluate the susceptibility to conventional antimicrobials and natural extracts of isolates that originated from cases of subclinical mastitis in dairy goats, as well as the biofilm production in interaction with the extract of Hymenaea martiana.
Location
This experiment was carried out in the Laboratory of Microbiology and Animal Immunology on the experimental farm of the Federal University of Vale do São Francisco – UNIVASF, Rodovia BR 407, km 12 – Lote 543 – Projeto de Irrigação Senador Nilo Coelho, s/nº “C1”, Petrolina-PE.
Obtaining the ethanol extract of Hymenaea martiana Hayne
The plant material was collected in the district of Petrolina-PE and identified in the Reference Center for the Recovery of Degraded Areas (CRAD) of the Universidade Federal do Vale do São Francisco. One plant specimen (21,868) was deposited in the Herbarium of the Vale do São Francisco (HVASF). The crude and solvent fractions of the plant bark were prepared as previously described (Silva et al., 2012).
Phytochemical screening
For the phytochemical screening of the ethanolic crude extract (CEE), chromatography analysis was conducted using thin-layer chromatography on plates of silica gel 60, with aluminum support and fluorescence F254 in different solvent systems. This analysis was performed following previously described methodology (Wagner and Bladt, 1996) to investigate alkaloids, anthracene derivatives, coumarins, flavonoids, tannins, lignans, monoterpenes, diterpenes, naphthoquinones, triterpenes and steroids. HPLC-DAD analysis was performed to identify the profile of the phenolic compounds. The results obtained using HPLC-DAD indicated the presence of three compounds (flavonoid derivatives) (Peixoto et al., 2015).
Bacterial samples
In total, 33 isolates from the Laboratory of Microbiology and Animal Immunology were used. These isolates were first identified as belonging to the genus Staphylococcus based on their biochemical characteristics (Gram, catalase, coagulase, DNase and sugar fermentation). These isolates came from goats with subclinical mastitis in the district of Valente-BA.
All of the isolates were tested for the presence of the nuc and rdr genes (Kateete et al., 2010; Shome et al., 2012), which would classify the microorganism as part of the Staphylococcus aureus or Staphylococcus epidermidis species, respectively.
While investigating the nuc gene, DNA that was extracted from the bacterial isolates was used as template, with 4 ml of the suspension added to 11 ml of the mix containing 2 mM MgCl2, 0.4 pmol of the primers, 0.4 mM deoxyribonucleotides, 1× enzyme buffer and 1.5 U of Taq DNA polymerase. The amplification cycles of the nuc gene were conducted with a number of modifications: initial denaturation at 94°C for 5 min; 37 cycles, each at 94°C, for 1 min; hybridization of the primer at 55°C for 30 s; extension at 72°C for 1 min; and a final extension at 72°C for 7 min (Kateete et al., 2010).
Polymerase chain reaction (PCR) was used to study the rdr gene with a number of modifications: 5 ml of the DNA suspension was added to 20 ml of the mix containing 1.5 mM MgCl2, 0.5 pmol of the primers, 0.4 mM deoxyribonucleotides, 1× enzyme buffer and 1.5 U of Taq DNA polymerase. The amplification cycles of the rdr gene involved an initial denaturation at 94°C for 5 min; 30 cycles, each at 94°C, for 30 s; hybridization of the primer at 60°C for 30 s; extension at 72°C for 45 s; and a final extension at 72°C for 10 min (Shome et al., 2012).
The results of the PCR were confirmed with a 1.5% agarose gel that was stained with ethidium bromide (1.0 mg mL-1) and an UV transilluminator was used to see the bands. Table 1 describes the primers that were used.
Phenotypic test of the sensitivity to antimicrobials
The sensitivity profile of the microorganisms was determined using the Kirby-Bauer disc diffusion method. The isolates were seeded in Mueller Hinton broth and incubated at 37ºC until a turbidity of 0.5 on the McFarland scale was obtained. A swab was used to seed the isolates in Petri dishes containing a medium of Mueller Hinton agar culture. Subsequently, discs that were impregnated with the following drugs were applied: ampicillin (10 μg), doxycycline (30 μg), lincomycin (2 μg), erythromycin (15 μg), gentamicin (10 μg), rifampicin (30 μg), cephalothin (30 μg), amoxicillin (10 μg), nalidixic acid (30 μg) and oxacillin (1 μg). The plates were incubated in an oven for 24 h at 37ºC. The diameter of the zone of inhibition was measured to determine the sensitivity profile of the isolates (CLSI, 2019). Afterwards, the multiple antibiotic resistance index (MARI) was calculated.
Molecular analysis of the genetic potential for resistance to antimicrobials
The aim of the PCR was to assess the presence of genes of resistance. To achieve this aim, it was necessary to extract DNA from the isolates in advance. The DNA was extracted and purified based on modified versions of previously described protocols (Ausubel et al., 1989; Aldous et al., 2005). A loopful culture was placed in 300 µl of TE (Tris-EDTA), and the samples were vortexed for homogenization. Subsequently, 70 µl of 10% SDS was added, and the samples were homogenized once again. In the next step, 100 µl of 5 M NaCl2 and 80 µl of CTAB/NaCl were added. The samples were incubated at 65°C for 10 min. Then, 700 µl of chloroform/isoamyl alcohol (24:1) was added, and the samples were homogenized by inversion. The samples were then centrifuged at 11,750 g for 5 min. The first phase was transferred to another tube, and 450 µl of isopropanol was added. The tubes were inverted and left on ice for 20 min. Thereafter, the tubes were centrifuged at 11,750 g for 15 min; subsequently, the supernatant was discarded, and 500 µl of 70% ethanol was added. Next, the samples were centrifuged at 11,750 g for 10 min. The supernatant was then discarded, and the microtubes were inverted to facilitate drying. Finally, the samples were suspended in 50 µl of TE (pH 8.0) and left at 65°C for 10 min. Once completed, the samples were stored at -20°C.
The genetic potential for resistance to antimicrobials was analyzed by amplifying the mecA and blaZ genes for resistance to methicillin (oxacillin) and beta-lactam, respectively. The DNA that was extracted from the bacterial isolates was used as a template to amplify a fragment of 214 bp corresponding to the mecA gene and a fragment of 517 bp corresponding to the blaZ gene. In total, 4 ml of this suspension was added to 11 ml of a mix containing 2 mM MgCl2, 0.4 mM primers, 0.4 mM deoxyribonucleotides, 1x enzyme buffer and 1.5 U of Taq DNA polymerase. The amplification reaction of the mecA gene involved an initial denaturation at 94°C for 1 min, 15 cycles of denaturation at 94°C for 30 s, 68°C for 30 s, 72°C for 30 s, an additional 20 cycles of 30 s at 94°C, 30 s at 60°C, 30 s at 72°C and a final extension at 72°C for 2 min (Murakami et al., 1991). The amplification reaction of the blaZ gene involved an initial denaturation at 94°C for 4 min, 30 cycles at 94°C for 1 min, 50.5ºC for 30 s, 72°C for 30 s, and a final extension at 72°C for 5 min (Sawant et al., 2009). Table 1 describes the primers that were used.
Molecular analysis of the genetic potential for the formation of biofilm (icaD, icaA and bap)
The DNA that was extracted from the bacterial isolates was used as a template. For the icaD gene, 2 ml of this suspension was added to 13 ml of the mix containing 1.33 mM MgCl2, 1 mM primers, 0.2 mM deoxyribonucleotides, 1x enzyme buffer and 1.5 U of Taq DNA polymerase. The reaction was kept in the thermocycler and submitted to an initial denaturation at 94°C for 2 min, followed by 30 cycles of 45 s at 92°C, 49.8°C for 45 s and 1 min at 72°C, with a final extension at 72°C for 7 min (Vasudevan et al., 2003).
For the icaA gene, 5 µL of the suspension (template) was added to 20 µL of the mix containing 2.5 mM MgCl2, 1 pmol of the primers, 0.28 mM deoxyribonucleotides, 1× enzyme buffer and 2.5 U of Taq DNA polymerase. The amplification reaction of the icaA gene was conducted as previously described (Vasudevan et al., 2003) with a number of modifications: initial denaturation at 94°C for 2 min, 30 cycles of 45 s at 92°C, 58.6°C for 45 s and 1 min at 72°C, with a final extension at 72°C for 7 min.
To study the bap gene, 5 µL of the suspension (template) was added to 20 µL of the mix containing 2.0 mM MgCl2, 0.4 pmol of the primers, 0.4 mM deoxyribonucleotides, 1× enzyme buffer and 2.5 U of Taq DNA polymerase. The amplification reaction of the bap gene was conducted with a number of modifications: initial denaturation at 94°C for 2 min, followed by 35 cycles of 45 s at 9°C, 56.5°C for 45 s and 50 s at 72°C, with a final extension at 72°C for 5 min (Cucarella et al., 2001). Table 1 describes the primers that were used.
Phenotypic test to detect the presence of biofilm
The phenotype of the isolates, in relation to the formation of biofilm, was characterized using the plate adherence test. The colonies that were isolated were inoculated in 3 mL of Tryptone Soya Broth (TSB) with glucose (0.25%) and incubated at 37°C for 24 h. Next, 200 µl was inoculated in micro-dilution plates and incubated again at 37°C for 24 h. After this period had elapsed, the plates were washed three times with 200 µl of distilled water and left to dry at room temperature. The plates were stained with 100 µl of crystal violet 0.25% for 2 to 3 min at room temperature and washed three more times with distilled water. To dissolve the dye, 200 µl of ethyl alcohol was used (80:20). The absorbance was measured using an Elisa Easy® micro-plate reader and a filter of 620 nm. All of the samples were analyzed in triplicate, as were the positive and negative controls. A strain of S. aureus ATCC 25923, which is genotypically characterized as a biofilm producer, was used as the positive control while a strain of S. epidermidis was used as the negative control. It was possible to determine biofilm production using the following classification: no biofilm production (optical density (OD) sample ≤ OD negative control); weak biofilm production (OD negative control < OD sample ≤ 2 OD negative control); moderate biofilm production (OD negative control < OD sample ≤4 OD negative control) and strong biofilm production (OD sample <4 OD negative control) (Merino et al., 2009).
Test of sensitivity to the extract
The crude ethanolic extract of H. martiana and the fractions that were obtained in ethyl acetate, hexane and chloroform were used in this test. First, 0.25 g of each extract was weighed and diluted in 10 mL of 1% DMSO, obtaining a stock solution with a concentration of 25 mg mL-1. The following concentrations were used: 12,500; 6,250; 3,125; 1,562.5; 781.2; 390.6; 195.3; and 97.6 µg/mL.
To prepare the inoculum, colonies that were obtained in Mueller-Hinton agar were used to create a bacterial suspension with turbidity of 0.5 on the McFarland scale. Of this suspension, 10 µL was inoculated in the wells of micro-plates containing the dilution of ethanolic extract. The material was incubated at 37°C for 24 h under aerobic conditions. The minimum inhibitory concentration (MIC) was determined to confirm the lowest concentration of the extract that was capable of inhibiting bacterial growth. When no bacterial growth was visible, one aliquot of 10 µl was withdrawn, seeded on the MH agar surface and incubated for 48 h at 37ºC. The following procedure was followed to determine the minimal bactericidal concentration (MBC): one aliquot of 10 μL was withdrawn from the wells without visible bacterial growth and seeded on the surface of the Mueller-Hinton agar. After 48 h of incubation at 35°C, the MBC was defined as the lowest concentration of the ethanolic extract that was required to cause the death of the inoculum. All of the trials were performed in triplicate.
Interaction of the extract with biofilm in the formation and consolidation of biofilm
Eleven (n=11) isolates that were identified as S. aureus were used to perform these tests. The biofilm in micro-plates was formed by incubating 100 µl of the bacterial suspension for 24 h at 37°C. Subsequently, the wells were washed three times with distilled water, and 100 µl of the extract was added, as described: 1/2, 1/4 and 1/8 of CBM were used for the tests. The optical density (OD) was determined immediately after the extract was added (0 h) and 24 h later.
The effect of the extract on the consolidated biofilm was defined by the following equation: OD0 h mean /OD24 h mean × 100 (Nostro et al., 2007).
Based on the MBC results that were obtained, three different concentrations (1/2, 1/4 and 1/8 of CBM) were used with biofilm in formation. This trial was conducted in micro-plates. The bacterial inocula were cultivated in 10 mL of TSB (1% glucose) for 24 h at 37°C. In total, 100 µl was added to the well plates, while 100 µl of vegetable extract and 100 µl of the culture media were previously added to the controls. After 24 h of incubation at 37°C, the plates were submitted to Gentian violet staining (Merino et al., 2009). The effectiveness of the extract in interfering with the formation of biofilm was defined by the following equation:
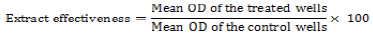
(Nostro et al., 2007).
Scanning electron microscopy
To assess the effect of the H. martiana extract on biofilm formation, an isolate of S. aureus that had been classified as a strong producer of biofilm was used to test for plate adherence. The control sample was inoculated in TSB broth. The same isolate was also inoculated in TSB broth containing the plant extract. Both were incubated at 37°C for 24 h. After the inoculum had been withdrawn from the suspension, it was inoculated in cover slips, which were then washed with sterilized saline solution for one minute. The slips were then fixed in glutaraldehyde (1%) for a period of 12 h. After fixation, the slides were immersed in increasing concentrations of ethanol (50, 70, 80, 95 and 100%), with 20 min between each alteration. After dehydration, the samples were immersed in acetone 100% and subjected to gold coating (Freitas et al., 2010). The fragments were studied using the TM-1000 Hitachi scanning electron microscope.
Statistical analysis
Descriptive statistics were used, including the distribution of relative and absolute frequencies, for the microbiological and molecular findings. Friedman’s analysis of variance was used to compare the MBC values in the three fractions of the extract. The Mann-Whitney test was used to assess the differences between the MBC values that were obtained for strong and moderate biofilm producers and for weak or non-producing isolates. The same test was used to compare the results of the MBC values between the isolates that were positive and negative for the blaZ gene (Thrusfield, 2007).
Phytochemical screening
The phytochemical screening of the CEE confirmed the presence of phenolic substances, flavonoids, steroids, and terpenoids (Table 2).
Bacterial samples
The molecular tests performed for the investigation of the nuc and rdr genes demonstrated that 97% (32/33) were positive for the nuc gene, being identified as S. aureus. Only one isolate was positive in the investigation of the rdr gene, indicating S. epidermidis.
Phenotypic test for sensitivity to antimicrobials
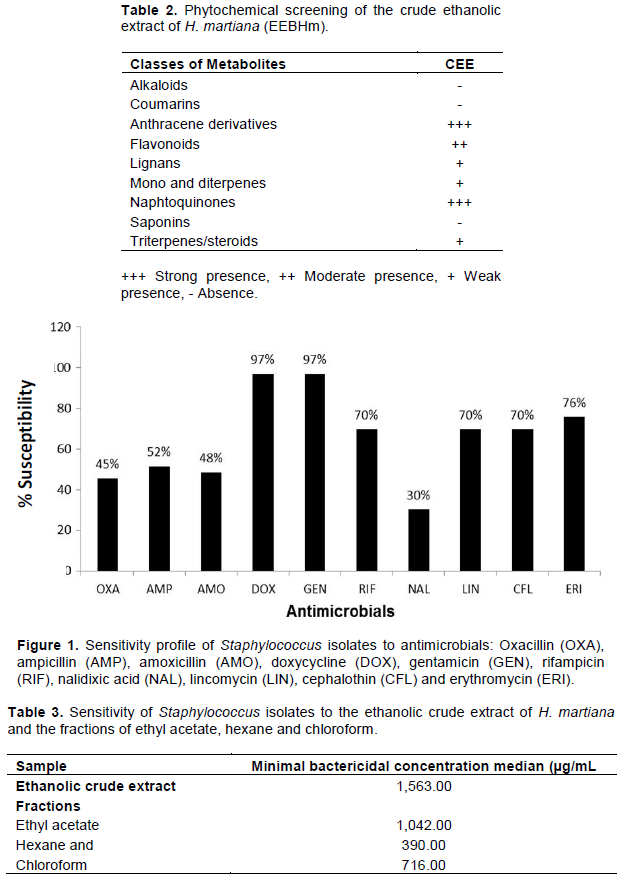
The sensitivity profile to antimicrobials of the evaluated isolates presented variation, which can be seen in Figure 1. In the susceptibility testing to antimicrobials, those with the highest percentage of in vitro sensitivity were gentamicin (97%) and doxycycline (97%), followed by erythromycin, with 76%, rifampicin, lincomycin, and cephalothin, with 70%, and with no great variation between oxacillin, ampicillin, and amoxicillin, with 45, 52, and 48%, respectively. The drug with the lowest sensitivity percentage was the nalidixic acid, with 30%. Among the evaluated samples, the IRMA varied from 0 to 0.8, of which 51.5% (17 isolates) presented this index above 0.2; that is, were resistant to two or more antimicrobials.
Molecular analysis of the genetic potential for resistance to antimicrobials
The resistance to oxacillin was observed in 18 (54.5%) isolates; however, no amplification by PCR of the mecA gene was observed. Out of the 33 isolates evaluated, 11 (33.3%) were positive for the blaZ gene.
Molecular analysis of the genetic potential for biofilm formation
Out of the 33 evaluated samples, nine (27.2%), three (10.0%), and four (12.9%) presented the icaD, icaA, and bap genes, respectively. Only two samples were positive at the same time for the icaD and icaA genes.
Phenotypic test to detect the presence of biofilm
Out of the 33 evaluated samples, 1 (3%) presented strong biofilm production, 9 (27.2%) presented moderate biofilm production, 15 (45.4%) presented weak production, and 8 (24.2%) were classified as non-biofilm producers.
Test of sensitivity to the extract
In the in vitro sensitivity tests to the crude ethanolic extract of H. martiana (EEBHm), the means obtained for the MBC of the crude ethanolic extract and its fractions are presented in Table 3, and no significant difference was observed (p >0.05). For the crude ethanolic extract, a median of 2864 µg/mL was observed among the isolates classified as moderate or strong biofilm producers. On the other hand, among the isolates classified as negative or weak biofilm producers, the mean MBC obtained for the crude extract of H. martiana was equal to 1563 µg/mL (p=0.07). Among the 11 positive isolates for the blaZ gene, a median of 781 µg/mL was observed. On the other hand, among the negative isolates for the presence of this gene, the median was equal to 2604 µg/mL, although without a significant statistical difference (p=0.14).
Interaction between the extract and biofilm in the formation and consolidation of biofilm
It was observed that, after the addition of the extract, 81.8, 90.0, and 81.83% of the isolates presented a reduction in biofilm production in the ethyl acetate, hexane, and chloroform fractions, respectively. Regarding the observation of biofilm after its establishment, no reduction in production was observed in any of the isolates (Table 4).
Scanning electron microscopy
In the control sample, which had no contact with the plant extract, a wide extracellular matrix production was observed. On the other hand, in the sample that remained in contact with the extract for 24 h, no biofilm production was observed (Figure 2).
Hymenaea is a genus that is distributed throughout tropical America, from Mexico to Paraguay, with one species located in coastal East Africa (Mackinder, 2005). The phytochemical screening of the CEE confirmed the presence of phenolic substances, flavonoids, steroids and terpenoids. Flavonoids are secondary metabolites and can be found in moderate concentrations in the extract that was used in the present study. The phytochemical screening of the crude ethanolic extract confirmed the presence of phenolic substances, flavonoids, steroids and terpenoids, as displayed in Peixoto et al. (2015).
In the test of susceptibility to antimicrobials, it is clear that there were only small variations between beta-lactams, with a similar resistance profile to the drugs in this group. Under similar experimental conditions, researchers recorded strong resistance among isolates from cases of subclinical mastitis in small ruminants (França et al., 2012). It is notable that a number of isolates exhibited resistance to oxacillin. A previous study expressed concern about the increasing occurrence of multi-drug resistance among microorganisms isolated from small ruminants with mastitis (Silva et al., 2004). Regarding the drugs in the group known as aminoglycosides, a high number of sensitive isolates were recorded, corroborating the findings of other studies (França et al., 2012; Kumar et al., 2009).
The mecA gene was not detected in the samples that were tested in the present study. A previous study reported that isolates that are resistant to oxacillin in several phenotypic tests cannot exhibit this gene given that the phenotype that produces beta-lactams, including class D oxacillinases, is resistant to penicillin (Soares et al., 2008). However, the assessments of the blaZ gene confirmed a positivity of 33.3% (n=11). Of this total, four (4) isolates were resistant to the beta-lactams that were tested. The results indicate the codified production of beta-lactam by the blaZ gene, a mechanism that has been reported in dairy cattle from other countries (Olsen et al., 2006; Taponen and Pyörälä, 2009). Although antimicrobial therapy focusing on mastitis in the sampled herds is not very common, different drugs are used for other infections in small ruminants. The generalized use of certain antimicrobials for clinical treatment could be associated with a high rate of resistance.
An analysis of the MARI scores indicated that more than half of the isolates exhibited indices that were greater than 0.2. This pattern of multi-resistance to antimicrobials has increased due to the exaggerated use of these drugs (Ferreira et al., 2006). The indices of resistance that were recorded in the present study could have been caused by the incorrect management methods that were used on the properties. Consequently, this pattern provides the lowest number of drugs available for use in the therapy of mastitis.
Among the isolates that were positive for the blaZ gene (n=11), nine exhibited mean MBC values less than 2,604 µg/mL, suggesting antimicrobial activity in the extract studied against the isolates that were carrying genes of resistance. However, it is essential to perform further studies to elucidate the mechanisms of the antibacterial action of the extract.
A molecular analysis of biofilm production confirmed the presence of the icaD, icaA and bap genes, although their frequencies were low. Only one of the isolates exhibited the concomitant presence of the icaD and icaA genes, which are responsible for the synthesis of biofilm. The genetic mechanisms that determine the production of biofilm are complex and involve other genes that were not investigated in the present study, including the IS257 gene (Tormo et al., 2005). A previous study demonstrated that molecular techniques to identify the ica genes that codify the synthesis of biofilm are important tools in the identification of virulent strains (Arciola et al., 2002). The implication of biofilm in chronic bacterial infections in many species has unleashed a growing interest in the characterization of the genes involved in its formation (Vautor et al., 2008). The bap gene was recently identified as the gene that is responsible for the codification of a protein associated with biofilm. However, currently, this gene has only been found in a small proportion of S. aureus strains for bovine mastitis in Spain (Cucarella et al., 2001). A study in France investigated the presence of the bap gene in S. aureus isolates from different species and regions using PCR and found negative results for all of the isolates, suggesting that the prevalence of this gene among S. aureus isolates must be very low (Vautor et al., 2008).
Among the strains that were classified as negative or weak producers of biofilm in the present study, the best antimicrobial activity was recorded for the crude ethanolic extract of H. martiana. Furthermore, the production of biofilm decreased after the addition of the plant extract, thereby demonstrating its potential in terms of affecting the production of this extracellular matrix. However, no reduction was recorded when the extract was added after the consolidation of biofilm. The reductions in biofilm production was also confirmed by scanning electron microscopy and suggest in vivo biological activity among the compounds present in the natural extract, mainly flavonoid derivatives. Biofilm affects the activity of antimicrobials due to low penetration and to the phenotypically protected state of the bacteria that compose the structure (Jacques et al., 2010; Altieri et al., 2013). Previous studies have listed the following strategies to reduce biofilm: prevention of microbial fixation; prevention of microbial growth; interruption of cell-cell communication; inhibition of the synthesis of the matrix; and disintegration of the matrix of biofilm. The inhibition of biofilm formation depends on factors that are associated with the enzymatic inhibition of proteases and quorum-sensing mechanisms (González-Ortiz et al., 2014). Because we are dealing with a natural antimicrobial, the reduction in the production of biofilm after interaction between the isolate and the H. martiana extract requires further research that will enable a better understanding of the action of this extract. A previous study revealed that the use of inhibitors could drastically alter the treatment of many infectious diseases (Cegelski et al., 2008). The basic strategy to discover biofilm inhibitors involves the screening of chemical compounds during trials that assess the effects of drugs or extracts on the formation of biofilm (Landini et al., 2010). The same study stated that till date, studies have focused on “quorum sensing” inhibitors, compounds that affect the metabolism of the c-di-GMP molecule or inhibitors of the biosynthesis of DNA and nucleotides.
Although studies of alternative therapy for diseases in veterinary medicine are common, they are characterized by discontinuity, with very small numbers of studies about the same plant. Thus, the minimal data that are required to use plants in veterinary medicine are not always obtained, as pre-clinical and clinical trials are not carried out.