ABSTRACT
Fungi play important roles, as both decomposers and plant symbionts in mangrove ecosystems. Their ability to survive extreme environmental conditions makes them potential rich sources for recovery of novel bioactive compounds. In this study, biotechnological potential of fungi recovered from the rhizospheres of four mangrove species (Sonneratia alba, Rhizophora mucronata, Ceriops tagal and Avicennia marina) was explored. Rhizospheric sediment samples of the mangroves were collected using standard protocols and different media used for isolation of fungi. A total of 33 fungal isolates were recovered and characterized based on morphological, physiological and Internal transcribed spacer (ITS) gene region analysis. The disk diffusion method was used to evaluate the antibacterial activity of the isolates. Morphologically, the isolates showed different characteristics with regard to color, margin and elevation. Physiologically, all the isolates were able to grow at different pH ranges, ranging from pH 4 to pH 12 and at different temperatures from 20 to 30°C and tolerated sodium chloride up to 7%. Phylogenetic analysis grouped the isolates into two phyla: Ascomycota and Basidiomycota affiliated to different genera; Penicillium, Aspergillus, Talaromyces, Diatrypella, Thielavia, Hypocreales, Paracremonium, Geosmithia, Peniophora, Massarina and Fulvifomes. A total of 17 representative isolates produced inhibition zones against three pathogenic bacterial strains. The findings demonstrate that the tropical mangroves rhizospheres are a rich source of fungi with antibacterial compounds and traits relevant for biotechnological application.
Key words: Antimicrobial activity, tropical marine fungi, Mangrove sediments, Internal transcribed spacer (ITS).
Mangrove forests are considered a dynamic ecotone between terrestrial and marine habitats (Thatoi et al., 2013). They occupy about 181,000 km2 worldwide (Jusoff, 2013). The mangrove ecosystems cover a quarter of the world’s tropical coastlines and are major drivers for transferring organic matter from land to oceans (Simões et al., 2015). They form a niche with a wide spectrum of microbial diversity with bacteria and fungi constituting 91% of the total biomass of mangrove ecosystems (Simões et al., 2015). Fungi are important soil component as both decomposers and plant symbionts (Liu et al., 2015). They play major roles in ecological and biogeochemical processes, contributing significantly to the degradation of mangrove-derived organic matter (Ghizelini et al., 2012). Similar to other ecosystems, fungi are believed to play an important role in carbon and nitrogen recycling in mangrove ecosystems (Reef et al., 2010). Other studies have suggested that mangrove fungi serve as a source of biologically active natural products (Wu et al., 2016).
Although a considerable number of fungi have been isolated from the mangrove ecosystems from time to time, they were first reported from mangrove roots in Australia (Cribb and Cribb, 1956). Since then, there is still a need for information on mangrove- associated fungi. This is important in order to gain new insight into the fungal communities in mangrove ecosystems, since most studies conducted in these ecosystems have mainly concentrated on bacterial diversity and less attention is given to fungal communities (Simões et al., 2015). Additionally, there is alarming increase of emerging and re-emerging diseases. This is due to newly discovered and constantly evolving pathogens. Considering the potential benefits of fungi, the search for natural biologically active compounds from fungi against pandemics and infectious agents is necessary.
The potential for the recovery of anticancer, antioxidant and antimicrobial compounds makes the prospecting for mangrove fungi important (Thatoi et al., 2013). Mangrove ecosystems are hotspots for morphologically and physiologically diverse fungal groups, considering that the species have adapted to the extreme environmental conditions (hypoxic, fluctuating pH, tidal pressures, high salinity, etc.). Anticancer / antioxidant biomolecules (Bhimba et al., 2012; Prathyusha et al., 2018), and novel enzymes with potential for biotechnological applications have been recovered from fungi isolated from mangroves (Wu et al., 2018). Though an impressive panoply of novel biomolecules have been recovered from mangrove associated fungi, much of the focus has been on endophytic fungi (Guerrero et al., 2018; Kuzhalvaymani et al., 2019), while free-living mangrove fungi remain highly underexplored. The recent discovery of five novel funicone derivatives from Penicillium pinophilum SCAU037 byHe et al. (2019)demonstrates the biotechnological potential of free-living fungi adapted to mangrove sediments. Having in mind that plant species and geographical location can influence the diversity of microbes (Wu et al., 2016), this study was designed with an aim of isolating, characterizing and exploring the biotechnological potential of fungi from the rhizospheres of four mangrove species found in Mida creek (a polluted mangrove habitat) and Gazi Bay (a pristine mangrove site).
Site description
This study investigated two mangrove sites (Mida creek and Gazi bay) in Kenya (Figure 1) for the presence of potential biotechnologically relevant fungal species. Gazi bay is located in Kwale County (04°44′S 039°51′E), on the South Coast of Kenya and approximately 55 km south of Mombasa. The Chale peninsula to the East and a fringing coral reef in the South protects the Bay from strong sea waves. The climate is mostly humid and hot and the average annual humidity and temperature are about 95% and 28°C respectively. The total annual precipitation ranges from 1000 to 1600 mm (Lang’at, 2008). Mida Creek is located in Kilifi County (03°34′S, 039°96′E), approximately 25 km South of Malindi town and about 88 km North of Mombasa (Lang’at, 2008). Monthly temperature ranges between 23 and 27°C, with monthly maximum at 34°C in the hottest months and minimum at 20°C in the coldest months. The total annual precipitation ranges from 1000 - 1600 mm (Lang’at, 2008).
Sample collection
Rhizospheric sediment samples were obtained from four mangrove species (Sonneratia alba, Rhizophora mucronata, Ceriops tagal and Avicennia marina) common to the two study sites. Sampling was done in May 2018 according to previously described methods (Wu et al., 2016). Four mangrove trees of each species at intervals of 10 m were selected. For each individual mangrove species, the rhizosphere sediments (~100g) were sampled vertically along the base of the plant at two depths (1-5 cm and 10-15cm), using a standardized core sampler (Giannopoulos et al., 2019). A total of 32 samples (4x4x2) per site were kept in sterile plastic bags, maintained on dry ice in a box before they were transported and stored at -20°C until further analyses was performed at the Laval University, Quebec City, QC, Canada.
Physicochemical analysis of sediment samples
Nutrient analyses of the soil samples for nitrogen, carbon, phosphorus, potassium, calcium, magnesium and sodium were conducted according to standard methods (Brupbacher et al., 1968). Determination of pH and electrical conductivity was done using the calcium chloride method at a ratio of 1:2 using a digital Corning pH meter 140 (Corning Life Sciences, Massachusetts, USA) and electrical conductivity meter type CDM 2d radiometer (Radiometer, Copenhagen, Denmark), respectively.
Sample pre-processing and isolation of fungi
The sediment samples were air dried at room temperature (27±1°C) for seven days and sieved with a 2.5 mm sieve to remove larger particles such as stone and plant debris in order to obtain a consistent soil particle size for isolation using the soil dilution technique. The isolation of fungi from the sediment samples was performed by serial dilution method. About 0.1 g of the sediment sample was suspended in 1ml sterile distilled water in a sterile 1.5 ml Eppendorf tube and serially diluted to 10−3. One hundred ðœ‡L of the10−1, 10−2, and 10−3 suspensions were spread in triplicate onto isolation media. Dilutions were spread into three different types of isolation media, which included Potato dextrose agar (PDA), Czapek dox agar and Glucose, peptone, yeast extract agar (GPYA). All media were prepared according to the manufacturer’s instructions and supplemented with streptomycin (100 μg ml-1) to prevent any bacterial growth. All the plates were placed in a microbiological incubator at 28°C for 7 to 28 days. Daily follow up was done to observe any growth on the plates.
Morphological characterization of isolates
Morphological characterization of the fungal isolates was done by studying the colony characteristics of the fungal culture with regard to color on the upper and lower surface of the culture plate, margin and elevation (Kuzhalvaymani et al., 2019).
Physiological characterization of the fungal isolates
Growth behavior of the fungal isolates at different temperature, pH and sodium chloride concentration ranges were determined. For temperature, a 5 × 5 mm mycelia growth disc was inoculated on PDA media and incubated at 20, 30, 40 and 50°C for 7 days. The radial growth in mm for the colonies was measured in triplicate and recorded. For pH, the PDA media was adjusted to pH 4, 6, 8, 10
and 12 using 1M HCl and 1M NaOH solutions and poured into sterile plates. The agar plates were then inoculated with a 5× 5 mm mycelia growth slice of the isolates and incubated at 30°C for 7 days. Radial growth was measured (millimeters) and recorded. Sodium chloride tolerance test was done by supplementing the media with different concentrations of sodium chloride (5, 7, 9, 11 and 15%). For each case, a 5 × 5 mm mycelia growth slice was inoculated and plates incubated at 30° for 7 days before the radial growth was measured (mm) and recorded.
Extraction of secondary metabolites from the fungal isolates
Three pieces of 5×5 mm mycelia growth discs of selected fungal isolates were inoculated into 500-ml Erlenmeyer flasks containing 300 mL soybean casein broth and incubated at room temperature for fourteen days under stationary conditions. The broth culture was filtered to separate the filtrate and mycelia. The filtrate was then used for testing of antimicrobial activity.
Screening of fungal isolates for antimicrobial activity
Three standard pathogenic bacterial strains, a Gram-positive (Staphylococcus aureus ATCC 25923) and two Gram-negative (Escherichia coli ATCC 25922 and Pseudomonas aeruginosa ATCC 27853) were used for the determination of antibacterial activity of extracts of the fungal isolates. The disk diffusion method (Balouiri et al., 2016)was used to evaluate the antibacterial activity of extracts from selected fungal isolates. Mueller-Hinton agar plates were inoculated with 100 mL of a suspension of bacteria (S. aureus, E. coli, or P. aeruginosa), adjusted to equal the turbidity of 0.5 McFarland standard. Inoculated plates were allowed to air dry. Paper disks (MAST, UK) were prepared by soaking sterile paper disks (Ø=6 mm) with prepared fungal extracts and dried before they were applied to the surface of the inoculated agar plates. Prepared plates were incubated at 30°C for 24h then the diameter of each inhibition zone was measured using a Vernier caliper (Mitutoyo, Tokyo, Japan). All the tests were done in triplicates.
Molecular characterization of the fungal isolates
Genomic DNA (gDNA) extraction and polymerase chain reaction (PCR) amplification
Genomic DNA was extracted from pure fungal cultures using the DNeasy Plant kit (QIAGEN GmbH, Hilden, Germany) according to the manufacturer’s instructions. The extracted gDNA was quantified using NanoDrop 2000 spectrophotometer (Thermo Fisher Waltham, MA, USA), as a template for the amplification of the ITS gene region. A set of ITS1 (5’-TCCGTAGGTGAACCTGCGG-3’) and ITS4 (5’-TCCTCCGCTTATTGATATGC-3’) universal primers for fungi (White et al., 1990)were used. PCR was carried out in a 25 µl reaction volume; consisted of 2.5 ml 10× PCR buffer, 0.75 µl MgCl2, 0.5 µldNTPs, 0.5 µl of each primer, 0.2 µl platinum Taq (Invitrogen), 1µl of template DNA and 19.05µl of water. Amplification was performed with initial heating at 95°C for 30s followed by 30 cycles of denaturation at 95°C for 40s, annealing at 54°C for 50 s and extension at 72°C for 1 min and a final extension period at 72°C for 5 min using MJ Research PTC-225 Peltier Thermal Cycler 9 (MJ Research, Nevada, U.S.A.). Amplicons were confirmed by visualization on 1% ethidium bromide stained agarose gels in a gel documentation chamber. The PCR products were sequenced directly using the Sanger sequencing platform at the Institute for Systems and Integrative Biology of Laval University, Canada. The sequences from this study have been deposited in GenBank
(www.ncbi.nlm.nih.gov/Genbank/submit.html) under the accession numbers, MT543100 - MT543132.
Phylogenetic analysis
Sequences of the isolates were manually edited in chromas and checked for presence of artifacts or sequencing errors using Mallard software (Ashelford et al., 2006), an NCBI bioinformatic tool for detecting chimera sequences. A search for similar sequences using BLASTN (Altschul et al., 1990)was performed, and sequence alignment was performed using the CLUSTAL Omega program (http://www.clustal.org) against the nearest neighbors. A neighbor-joining tree of the aligned sequences was constructed (Saitou and Nei, 1987)using MEGA X software (Kumar et al., 2018). Evolutionary distances were computed using the Maximum Composite Likelihood method (Tamura et al., 2004). To obtain statistical support values for the branches, bootstrapping (Felsenstein, 1985)was conducted with 1000 replicates. All sites, including gaps in the sequence alignment, were excluded pairwise in the phylogenetic analysis. Using the resultant neighbor-joining tree, each isolate was assigned to the proper taxonomic group. The taxonomic assignment was conï¬rmed at a 95% conï¬dence level using the RDP Naïve Bayesian rRNA Classiï¬er Version 2.11 on the RDP website (Wang et al., 2007).
Statistical analyses
Data from physicochemical parameters was analysed using R v3.6.1 (Somanathan et al., 2004). A two-factor (sites and mangrove species differences) test of differences in physicochemical parameters was done by the non-parametric Kruskal-Wallis H test using the agricolae (de Mendiburu, 2020)R package. Post hoc test for mean separations was based on Fisher’s least significant difference.
Data from physiological analysis and antimicrobial activity was analyzed using ANOVA followed by Tukey HSD post-hoc test using SigmaPlot version 14.0 (Systat Software, San Jose, CA). Results were expressed as the mean ± SD. A p-value of ≤ 0.05 was considered statistically significant. All experiments were performed independently at least three times.
Physicochemical analysis of sediment samples
The pH and calcium were significantly higher (Kruskal-Wallis, p ≤ 0.05) in all mangrove plant species of Mida creek compared to Giza bay, which had significantly higher electrical conductivity (EC) and salinity values (Table 1). Besides the rhizosphere of R. mucronata, all other mangrove species rhizosphere in Gazi bay had significantly lower (p < 0.05) physicochemical properties compared to the species in Mida creek. The physicochemical parameters that were higher in the rhizosphere of R. mucronata in Gazi bay included potassium, sodium, phosphorus, total carbon, nitrogen, salinity and EC (Table 1).
Morphological characterization of isolates
A total of 33 fungal isolates were obtained from the mangrove sediments. Isolates were generally slow growers as they grew between 7-28 days. Morphological characterization was based on traditional macroscopic features of color, margin and elevation of the pure colonies. The isolates exhibited different colony characteristics with the majority of them having curled margins and raised elevation. The reverse color was mostly black, while the obverse was mainly grey.
Physiological characteristics of the isolates
All the isolates exhibited growth at different pH ranges, but individual isolates exhibited optimal growth at different pH ranges as shown in Figure 2a. For instance, isolates AVM110F1, CTG25F1, AVM110F2 and AVG410F1 had no significant growth at the tested pH ranges (p-value > 0.05). RMG110F1 had a significant growth at pH 4 and pH 10. Isolates RMG110F2 and RMG210F1 grew optimally at pH 8, while AVG35F1 grew optimally at pH 10 and pH 12. Isolate SAG35F1 had a significant growth at pH 6 and 12, while CTG45F1 grew significantly at pH 4, pH 6 and pH 10 (Figure 2a). Growth at different temperatures was only observed at 20 and 30°C for all the isolates (Figure 2b). No growth was observed at 40, 50 and 60°C; however, growth differences of the isolates were observed at both temperatures (20 and 30°C). There were insignificant growth changes at both temperatures for isolates CTG25F1, AVM110F2, RMG110F2 and AVG410F2. Isolates AVM110F1, RMG110F1, SAG35F1, RMG15F1, CTG45F1, RMG25F1 and SAG15F1 grew optimally at 20°C, while isolates RMG210F1, RMM15F1, AVG410F1 and AVM15F2 were observed to grow optimally at 30°C. At different NaCl concentrations, 29.4% of the tested isolates exhibited insignificant growth compared to 58.8% of the tested isolates, which had relatively growth changes with increase in NaCl concentration (Figure 2c). Isolates RMG110F2, RMM15F1, RMG15F1, RMG25F1 and SAG15F1 had significant growth at 5 and 7% NaCl (p-value ≤ 0.05).
Screening of isolates for antimicrobial activity
The result on antibacterial activity of the fungal isolates against some known bacterial strains (E. coli, P. aureginosa and S. aureus) is presented in Table 2. Extracts of 17 fungal isolates showed different inhibition zone diameters on all or any of the three pathogens. Some isolates exhibited significantly higher inhibition zone diameters (P-value ≤ 0.05) as indicated in Table 2. For instance, isolates RMG110F1, RMG210F1, AVG35F1, RMG35F1, AVG410F1, SAG35F1, RMG15F1, AVG410F2 and AVM15F2 produced significantly higher inhibition zone diameters against the tested bacterial strains. Notably, isolate AVG410F2 significantly inhibited the growth of all bacterial strains while isolates SAG35F1 and AVM15F2 had significantly higher inhibition zone diameters on E. coil only (p-value ≤ 0.05).
Molecular characterization of the fungal isolates
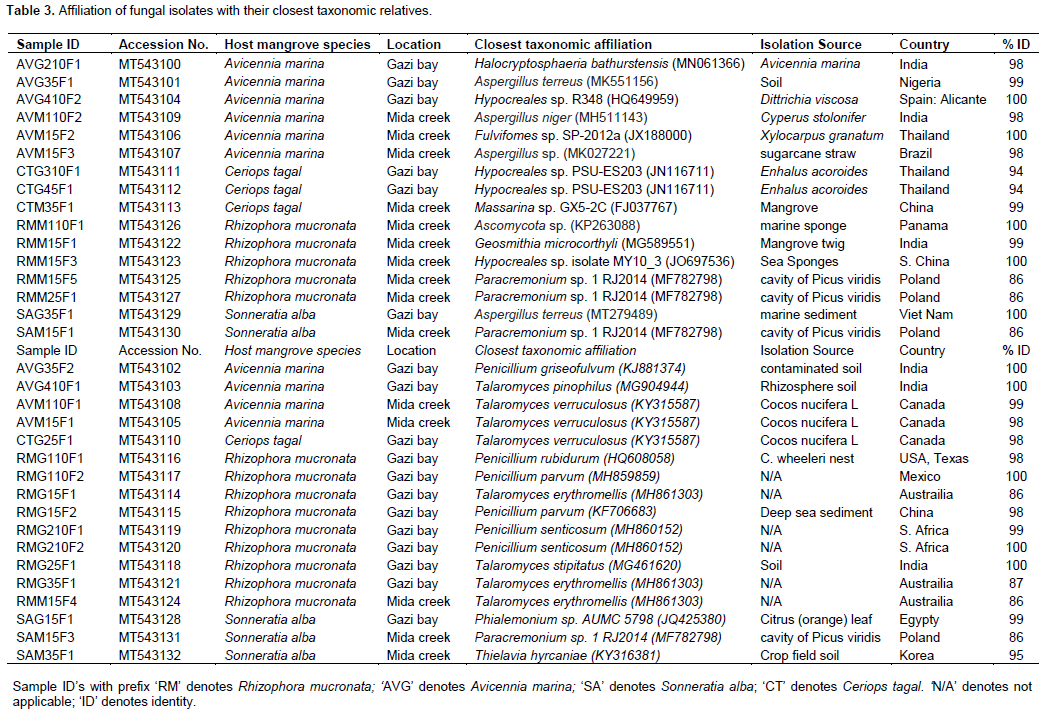
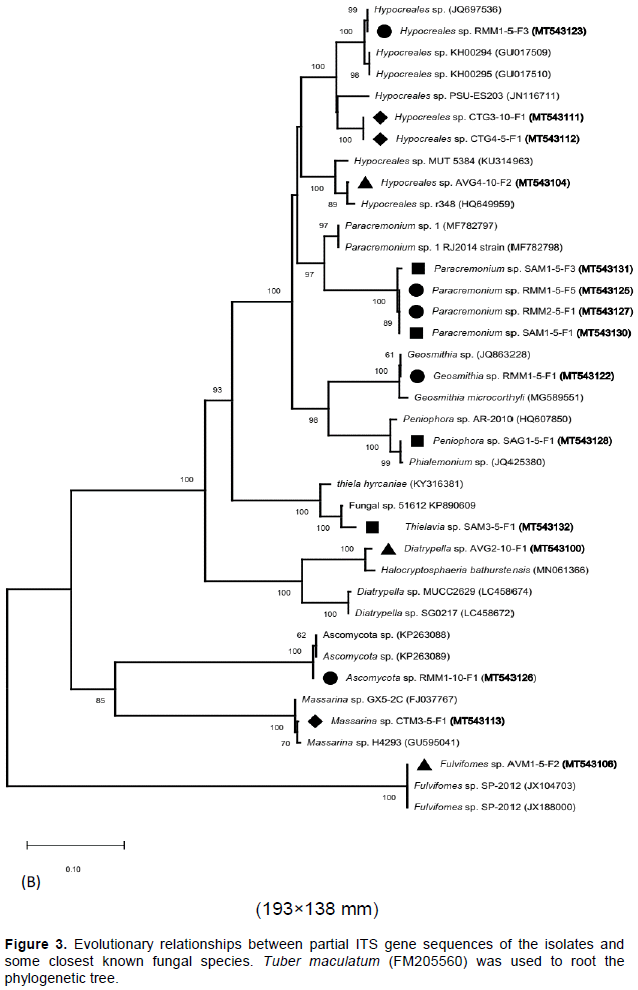
A total of 33 isolates were recovered from the rhizospheric sediment samples of four-mangrove species. About 43, 30, 15 and 12% of the total isolates were obtained from the rhizospheric sediments of Rhizophora mucronata, Avicennia marina, Sonneratia alba and Ceriops tagal, respectively as shown in Table 3. The isolates (with their accession numbers in parenthesis) in the inferred phylogenetic trees (Figure 3a, b) were phylogenetically diverse and affiliated with known genera (Penicillium, Aspergillus, Talaromyces, Diatrypella, Thielavia, Hypocreales, Paracremonium, Geosmithia, Peniophora, Massarina and Fulvifomes) belonging to two fungal phyla: Ascomycota (yeast and sac fungi) and Basidiomycota (club fungi). Comparison of the newly isolated ITS gene sequences to known sequences in the GenBank database using Blastn analysis indicated sequences similarities of between 85 to 100% with known sequences (Table 3).
Affiliation of ITS gene sequences of the isolates
Most of the isolates (~97%) were affiliated with several known fungal species from the phylum Ascomycota with >85% sequence identity (Table 3; Figure 3a, b). Eight isolates (RMG15F1 [MT543114], RMG35F1 [MT543121], RMM15F4 [MT543124], AVG410F1 [MT543103], RMG25F1 [MT543118], AVM110F1 [MT543108], AVM15F1 [MT543105] and CTG25F1 [MT543110]) had between 85-100% sequence identities with known members of the genus Talaromyces that together formed a single sub-cluster supported with a bootstrap value of 99% (Table 3; Figure 3a, b). Out of the eight isolates, four (RMG15F1 [MT543114], RMG35F1 [MT543121], RMM15F4 [MT543124] and RMG25F1 [MT543118]) were isolated from the rhizospheric sediments of R. mucronata, while three isolates (AVG410F1 [MT543103], AVM15F1 [MT543105] and AVM110F1 [MT543108]) were obtained from the rhizospheric sediments of A. marina. Isolate CTG25F1 (MT543110) was obtained from the rhizospheric sediments of C. tagal. Six isolates (AVG35F2 [MT543102], RMG15F2 [MT543115], RMG110F2 [MT543117], RMG110F1 [MT543116], RMG210F1 [MT543119] and RMG210F2 [MT543120]) were affiliated with members of the genus Penicillium [with >98% sequence identities] (Table 3) and together formed a minor sub-cluster supported with a bootstrap value of 90% (Figure 3a, b). All the six isolates were obtained from the rhizospheric sediments of R. mucronata except isolate AVG35F2 [MT543102], which was isolated from the rhizospheric sediments of A. marina.
Four isolates (RMM15F5 [MT543125], RMM25F1 [MT543127], SAM15F1 [MT543130] and SAM15F3 [MT543131]) had 86% sequence affiliation with Paracremonium sp. 1 RJ2014 (MF782798) and together formed a minor sub-cluster supported with a bootstrap value of 97% (Table 3; Figure 3a, b). Two of these isolates (RMM15F5 [MT543125] and RMM25F1 [MT543127]) were obtained from the rhizospheric sediments of R. mucronata, while the other two were isolated from the rhizospheric sediments of S. alba. Other four isolates (CTG45F1 [MT543112], AVG410F2 [MT543104], RMM15F3 [MT543123] and CTG310F1 [MT543111] were >94% affiliated with sequences of members from the genus Hypocreales and formed a minor sub-cluster supported by a bootstrap value of 100% (Table 3; Figure 3a, b). Out of these four isolates, two (CTG310F1 [MT543111] and CTG45F1 [MT543112]) were isolated from the rhizospheric sediments of C. tagal, while AVG410F2 [MT543104] and RMM15F3 [MT543123] were obtained from the rhizospheric sediments of A. marina and R. mucronata, respectively.
Four other isolates (AVM110F2 [MT543109], AVM15F3 [MT543107], AVG35F1 [MT543101] and SAG35F1 [MT543129]) were affiliated with members of the genus Aspergillus [with >98% sequence similarities] (Table 3). These isolates formed a single sub-cluster with several other members of the genus Aspergillus as indicated in the inferred phylogenetic tree (Figure 3a, b). Three of these isolates (AVM110F2 [MT543109], AVM15F3 [MT543107] and AVG35F1 [MT543101]) were obtained from the rhizospheric sediments of A. marina, while SAG35F1 [MT543129] was isolated from the rhizospheric sediments of S. alba. Isolate CTM35F1 [MT543113] had a 99% sequence identity with Massarina sp. GX5-2C (FJ037767) and formed a separate minor sub-cluster supported by a bootstrap value of 85%. In addition, this sub-cluster has members of the Ascomycota species together with isolate RMM110F1 [MT543126], which was obtained from the rhizospheric sediments of R. mucronata (Table 3; Figure 3a & b). Isolates (AVM15F2 [MT543106], AVG210F1 [MT543100], SAG15F1 [MT543128], SAM35F1 [MT543132] and RMM15F1 [MT543122]) were affiliated with sequences from different species with between 95-100% sequence identities. These isolates also formed minor sub-clusters as shown in the phylogenetic tree (Figure 3a, b) and were obtained from the rhizospheric sediments of A. marina, R. mucronata and S. alba (Table 3).

Fungal biodiversity studies in mangroves are essential for exploring diversity from not only the biogeographical perspective but also with the view of bioactive secondary metabolites biosynthesis capability (Zhou et al., 2016). In this study, 33 fungal isolates were obtained from the rhizospheric sediments of four mangrove species (R. mucronata, A. marina, S. alba and C. tagal) that are common along the Kenyan Coastline. Close observation of the distribution of the fungal isolates among the mangrove species showed that most isolates (43% of the total isolates) were recovered from the rhizospheric sediments of R. mucronata in both Gazi bay and Mida creek (Table 3). This observation could be explained by the variation in nutrients within the mangrove zonation, where areas occupied by R. mucronata species are richer in nutrients than other areas. Nitrogen, phosphorus, potassium, calcium, sodium, carbon and magnesium were among the nutrients higher in the rhizosphere of R. mucronata in both Gazi bay and Mida creek (Table 1).
Phylogenetic analysis positioned the isolates into three major clusters that were represented by eleven genera (Talaromyces, Aspergillus, Penicillium, Hypocreales, Paracremonium, Diatrypella, Thielavia, Geosmithia, Peniophora, Massarina and Fulvifomes) belonging to two fungi divisions namely Ascomycota (accounted for ~ 97% of all the isolates) and Basidiomycota (accounted for ~3% of the total isolates). It is worthy to note that most of these genera are saprophytic fungi and may be implicated in biodegradation of litter in the mangrove ecosystem (Hýsek and Brožová, 2001). Majority of the isolates (24%) formed a single major cluster supported by a bootstrap value of 99% and were affiliated with members from the genus Talaromyces [with ≥86% sequence identity] (Table 3). Initially, there were few reports about the occurrence of this genus in the mangrove ecosystems since it was classified under the genus Penicillium. But following its formal separation from the genus Penicillium, most researchers have detected this genus in mangrove ecosystems (Lima et al., 2018; Venkatachalam et al., 2019). Members of the genus Talaromyces form part of the major fungi adapted to the extreme conditions in mangrove ecosystems and have been reported to be a source of biotechnologically relevant bioactive molecules (Jie et al., 2016).
Interestingly, about 30% of the isolates were closely related to members of the genera Aspergillus and Penicillium [with >97% sequence identity]. Members from these genera are widely distributed including in the marine ecosystem (Zhou et al., 2018; Venkatachalam et al., 2019). For instance, isolate SAG35F1 [MT543129] had 100% similarity with Aspergillus terreus (MT279489), which was isolated from marine sediment in Vietnam, while isolate RMG15F2 [MT543115] had 98% sequence identity with Penicillium parvum (KF706683), which was recovered from deep sea sediment in China (Table 3). Moreover, members from these genera have been isolated from mangrove ecosystems by other researchers (Al-Shibli et al., 2019; Kuzhalvaymani et al., 2019). Other classified fungi with appreciable frequency of isolation were members of the genera Hypocreales and Paracremonium. Zhou et al. (2018)reported the occurrence of species from these genera in the mangrove environment. Members from other genera (Diatrypella, Thielavia, Geosmithia, Peniophora, Massarina and Fulvifomes) formed the least abundant species among the isolates. It is noteworthy that fungal genera like Geosmithia, Peniophora, Massarina and Fulvifomes have previously been reported in mangrove ecosystems (Osorio et al., 2014; Ameen et al., 2016; Guerrero et al., 2018). Members of the genus Diatrypella are cosmopolitan in distribution, and occur on a broad range of dead or dying wood of angiosperms, but there are fewer reports on their occurrence in the mangrove environment (Shang et al., 2017).
The physiological analysis revealed that all the isolates were able to grow at different pH ranges (from low to high pH values [pH 4 to 12]), but some had growth preference for either slightly acidic or alkaline environment (Figure 2a). Some isolates (Penicillium sp. [MT543117], Geosmithia sp. [MT543122], Talaromyces sp. [MT543114], Talaromyces sp. [MT543118] and Phialenonium sp. [MT543128]) grew at significantly lower NaCl concentrations (5 and 7%NaCl) compared to others, indicating difference in salinity levels preference. This is attributed to the pH fluctuations and high salinity levels in mangrove ecosystems (Venkatachalam et al. (2019); thus, these fungal isolates are adapted to such conditions. A related study by Venkatachalam et al. (2019)revealed that marine-derived genera Talaromyces and Penicillium, were the most promising for the production of compounds such as pigmented secondary metabolites, which can be applied in cosmeceutical or food industries. These authors also observed that the production of the pigmented metabolites was regulated by the concentration of salt in the growth media. There were insignificant growth differences at both 20 and 30°C for some isolates, but others (Isolates AVM110F1, RMG110F1, SAG35F1, RMG15F1, CTG45F1, RMG25F1 and SAG15F1) grew optimally at 20°C; while isolates RMG210F1, RMM15F1, AVG410F1 and AVM15F2 were observed to grow optimally at 30°C. This indicates that these isolates preferred normal marine environmental temperature ranges above which (40 to 60°C) growth was not observed.
Moreover, seventeen fungal isolates showed antibacterial activity against the pathogenic bacterial strains (E. coli, P. aeruginosa, and S. aureus). Isolates such as Penicillium sp. [MT543116], Penicillium sp. [MT543119], Aspergillus sp. [MT543101], Talaromyces sp. [MT543121], Talaromyces sp. [MT543103], Thielavia sp. [MT543132], Penicillium sp. [MT543115], Hypocreales sp. [MT543104] and Fulvifomes sp. [MT543106] exhibited significantly higher inhibition zone diameters against the bacterial strains (Table 2). This demonstrates that these fungal isolates have potential for formation of antimicrobial agents. Notably, isolate Hypocreales sp. [MT543104] significantly inhibited the growth of all tested bacterial strains, while isolates Thielavia sp. [MT543132] and Fulvifomes sp. [MT543106] had significantly higher inhibition zone for E. coil only (P ≤ 0.05). This could mean that these isolates produce different types of antibacterial compounds, which are either effective against a narrow range of bacteria or a broad range of bacteria. One of the notions is that this is a defense mechanism attributed to the intense competition between microbial species, which evolved to secrete substances with high antibacterial activity inhibiting the growth of other strains (Zhou et al., 2016). A recent effort at exploring mangroves rhizospheric fungi associated with the roots of Rhizophora stylosa, on the Techeng Isle in China led to the recovery of 5 novel funicone derivates from Penicillium pinophilum (He et al., 2019). Other studies (Bhimba et al., 2012; Lima et al., 2018)have also demonstrated that species from these isolated fungal genera have antimicrobial activity, which is consistent with these findings.
This study demonstrated that the distribution of culturable fungi in the mangrove ecosystem of Gazi bay and Mida creek is influenced by both plant species and differences in environmental conditions. The recovery of 17 fungal isolates with antimicrobial activity against known bacterial pathogens implies that mangroves rhizospheres are also hotspots for the isolation of fungi with antibacterial potential. Further analysis on the antibacterial activity of the isolated fungi will give more insights on the types of antibacterial compounds produced and their effectiveness as antimicrobial agents.
The authors have not declared any conflict of interests.
The authors appreciate the financial support provided by the Queen Elizabeth II Jubilee Scholarship to EMM and NSERC discovery grant to DPK and the International Foundation for Science (Ref No.: IFS Grant A/5719-1) to HMM. They are also grateful to NACOSTI, NEMA, KWS and KEPHIS for approving the research study and providing permits that facilitated field studies and shipment of samples to Canada. They appreciate the Centre for Forest Research and Institute for Systems and Integrative Biology of Laval University, Canada and Pwani University for their support during the project.
REFERENCES
|
Al-Shibli H, Dobretsov S, Al-Nabhani A, Maharachchikumbura SSN, Rethinasamy V, Al-Sadi AM (2019). Aspergillus terreus obtained from mangrove exhibits antagonistic activities against Pythium aphanidermatum-induced damping-off of cucumber. Peer Journal 10:1-16.
Crossref
|
|
|
|
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990). Basic local alignment search tool. Journal of Molecular Biology 215(3):403-410.
Crossref
|
|
|
|
|
Ameen F, Moslem M, Hadi S, Al-sabri AE (2016). Biodegradation of diesel fuel hydrocarbons by mangrove fungi from Red Sea Coast of Saudi Arabia. Saudi Journal of Biological Sciences 23(2):211-218.
Crossref
|
|
|
|
|
Ashelford KE, Chuzhanova NA, Fry JC, Jones AJ, Weightman AJ (2006). New screening software shows that most recent large 16S rRNA gene clone libraries contain chimeras. Applied Environmental Microbiology 72(9):5734-5741.
Crossref
|
|
|
|
|
Balouiri M, Sadiki M, Ibnsouda SK (2016). Methods for in vitro evaluating antimicrobial activity: A review. Journal of Pharmaceutical Analysis 6(2):71-79.
Crossref
|
|
|
|
|
Bhimba BV, Franco DAAD, Mathew JM, Jose GM, Joel EL, Thangaraj M (2012). Anticancer and antimicrobial activity of mangrove derived fungi Hypocrea lixii VB1. Chinese Journal of Natural Medicines 10(1):77-80.
Crossref
|
|
|
|
|
Brupbacher RH, Bonner WP, Sedberry Jr. J (1968). Analytical methods and procedures used in the soil testing laboratory 15. View
|
|
|
|
|
Cribb AB, Cribb JW (1956). Marine fungi from Queensland. II. Papers of the Department of Botany University of Queensland 3(12):97-105.
|
|
|
|
|
de Mendiburu F (2020). agricolae: Statistical Procedures for Agricultural Research (R package version 1.3-2).
|
|
|
|
|
Felsenstein J (1985). Confidence Limits on Phylogenies: An Approach Using the Bootstrap. Evolution 39(4):783.
Crossref
|
|
|
|
|
Ghizelini AM, Mendonça-Hagler LCS, Macrae A (2012). Microbial diversity in Brazilian mangrove sediments: a mini review. Brazilian Journal of Microbiology 43(4):1242-1254.
Crossref
|
|
|
|
|
Giannopoulos G, Lee DY, Neubauer SC, Brown BL, Franklin RB (2019). A simple and effective sampler to collect undisturbed cores from tidal marshes. bioRxiv 515825.
Crossref
|
|
|
|
|
Guerrero JJG, General MA, Serrano JE (2018). Culturable Foliar Fungal Endophytes of Mangrove Species in Bicol Region. Philippine Journal of Science 147:563-574.
|
|
|
|
|
He F, Li X, Yu JH, Zhang X, Nong X, Chen G, Zhu K, Wang YY, Bao J, Zhang H (2019). Secondary metabolites from the mangrove sediment-derived fungus Penicillium pinophilum SCAU037. Fitoterapia 136:104177.
Crossref
|
|
|
|
|
Hýsek J, Brožová J (2001). The role of some saprophytic micromycetes and fungus Micromucor ramannianus var. ramannianus in forest soil. Czech Mycology 53(2):61-171.
Crossref
|
|
|
|
|
Jenoh EM, Robert EMR, Lehmann I, Kioko E, Bosire JO, Ngisiange N, Dahdouh-Guebas F, Koedam N (2016). Wide ranging insect infestation of the pioneer mangrove Sonneratia alba by two insect species along the Kenyan coast. PLoS One 11:1-15.
Crossref
|
|
|
|
|
Jie MZ, Jiang LC, Di YSD, Crews P (2016). The Bioactive Secondary Metabolites from Talaromyces species. Natural Products Bioprospecting 6(1):1-24.
Crossref
|
|
|
|
|
Jusoff K (2013). Malaysian Mangrove Forests and their Significance to the Coastal Marine Environment. Polish Journal of Environmental Studies 22(4):979-1005.
|
|
|
|
|
Kumar S, Stecher G, Li M, Knyaz C, Tamura K (2018). MEGA X: Molecular evolutionary genetics analysis across computing platforms. Molecular Biology and Evolution 35(6):1547-1549.
Crossref
|
|
|
|
|
Kuzhalvaymani K, Elizabeth JL, Subha TS (2019). Isolation, Morphological characterization and Molecular identification of Endophytic fungi isolated from Mangroves. International Journal of Pharmacy and Biological Sciences 9(2):105-116.
|
|
|
|
|
Lang'at JKS (2008). Variability of mangrove forests along the Kenyan coast.
View
|
|
|
|
|
Lima MTNS, dos Santos LB, Bastos RW, Nicoli JR, Takahashi JA (2018). Antimicrobial activity and acetylcholinesterase inhibition by extracts from chromatin modulated fungi. Brazilian Journal of Microbiology 49(1):169-176.
Crossref
|
|
|
|
|
Liu P, Wang X, Li J, Qin W, Xiao C, Zhao X, Jiang H, Sui J, Sa R, Wang W, Liu X (2015). Pyrosequencing Reveals Fungal Communities in the Rhizosphere of Xinjiang Jujube. Biomed Research International 1-8.
Crossref
|
|
|
|
|
Osorio JA, Wing MJ, Roux J (2014). A review of factors associated with decline and death of mangroves, with particular reference to fungal pathogens. South African Journal of Botany 103:295-301.
Crossref
|
|
|
|
|
Prathyusha AMVN, Mohana SG, Bramhachari PV (2018). Chemical characterization and antioxidant properties of exopolysaccharides from mangrove filamentous fungi Fusarium equiseti ANP2. Biotechnology Reports 19.
Crossref
|
|
|
|
|
Reef R, Feller IC, Lovelock CE (2010). Nutrition of mangroves. Tree Physiology 30(9):1148-1160.
Crossref
|
|
|
|
|
Saitou N, Nei M (1987). The neighbor-joining method: a new method for reconstructing phylogenetic trees. Molecular Biology and Evolution 4(4):406-425.
|
|
|
|
|
Shang Q, Hyde KD, Phookamsak R (2017). Diatrypella tectonae and Peroneutypa mackenziei spp. nov. (Diatrypaceae) from northern Thailand. Mycological Progress 16:463-476.
Crossref
|
|
|
|
|
Simões MF, Antunes A, Ottoni CA, Amini MS, Alam I, Alzubaidy H, Mokhtar NA, Archer JAC, Bajic VB (2015). Soil and Rhizosphere Associated Fungi in Gray Mangroves (Avicennia marina) from the Red Sea - A Metagenomic Approach. Genomics, Proteomics and Bioinformatics 13(5):310-320.
Crossref
|
|
|
|
|
Somanathan H, Mali S, Borges RM, Shilton LA, Altringham JD, Compton SG, Whittaker RJ, Russo SE, Augspurger CK, Nakashima Y, Inoue E, Inoue-Murayama M, Abd Sukor JR, McConkey KR, Prasad S, Corlett RT, Campos-Arceiz A, Brodie JF, Rogers H, Rawat GS (2004). R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing. Oecologia 15:413-448.
|
|
|
|
|
Tamura K, Nei M, Kumar S (2004). Prospects for inferring very large phylogenies by using the neighbor-joining method. Proceedings of the National Academy of Sciences of United States of America 101(30):11030-11035.
Crossref
|
|
|
|
|
Thatoi H, Behera BC, Mishra RR (2013). Ecological role and biotechnological potential of mangrove fungi: a review. Mycology 4(1):54-71.
|
|
|
|
|
Venkatachalam M, Gérard L, Milhau C, Vinale F, Dufossé L, Fouillaud M (2019). Salinity and temperature influence growth and pigment production in the marine-derived fungal strain Talaromyces albobiverticillius. Microorganisms 7:1.
Crossref
|
|
|
|
|
Wang Q, Garrity GM, Tiedje JM, Cole JR (2007). Naïve Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Applied Environmental Microbiology 73(16):5261-5267.
Crossref
|
|
|
|
|
White TJ, Bruns T, Lee S, Taylor J (1990). Amplification and Direct Sequencing of Fungal Ribosomal RNA Genes for Phylogenetics. PCR Protocols and Methods pp. 315-322.
Crossref
|
|
|
|
|
Wu J, Qiu C, Ren Y, Yan R, Ye X, Wang G (2018). Novel salt-tolerant xylanase from a mangrove-isolated fungus phoma sp. MF13 and its application in Chinese steamed bread. American Chemical Society Omega 3(4):3708-3716.
Crossref
|
|
|
|
|
Wu P, Xiong X, Xu Z, Lu C, Cheng H, Lyu X, Zhang J, He W, Deng W, Lyu Y, Lou Q, Hong Y, Fang H (2016). Bacterial communities in the rhizospheres of three mangrove tree species from Beilun Estuary, China. PLoS One 11(10):1-13.
Crossref
|
|
|
|
|
Zhou J, Diao X, Wang T, Chen G, Lin Q, Yang X, Xu J (2018). Phylogenetic diversity and antioxidant activities of culturable fungal endophytes associated with the mangrove species Rhizophora stylosa and Rhizophora mucronata in the South China Sea. PLoS ONE 13(6):1-18.
Crossref
|
|
|
|
|
Zhou S, Wang M, Feng Q, Lin Y, Zhao H (2016). A study on biological activity of marine fungi from different habitats in coastal regions. SpringerPlus 5:1-8.
Crossref
|
|
