Review
ABSTRACT
Experimentally induced introgression and selection during domestication and maize (Zea mays L.) improvement involved selection of specific alleles at genes controlling morphological and agronomic traits, resulting in reduced genetic diversity relative to unselected genes. The plant breeder would have to extend crosses to the wild relatives to introduce novel alleles and diversify the genetic base of elite breeding materials. The use of maize wild relatives (Teosintes and Tripsacum) genes to improve maize performance is well established with important examples dating back more than 60 years. In fact, Teosintes and Tripsacum are known to possess genes conferring tolerance to several biotic and abiotic stress including chlorotic dwarf virus, downy mildew, Fusarium, Striga hermonthica, rootworms, drought and flooding. This review provides an overview of the application of these wild relatives and demonstrates their roles on the development of stress tolerant maize plants. It also highlights the use of Teosintes and Tripsacum to improve selected quantitative traits such as yield.
Key words: Maize (Zea mays L.), Teosintes, Tripsacum, stress tolerance, maize improvement.
INTRODUCTION
CHARACTERISTICS OF WILD ZEA SPECIES
PESTS AND DISEASE RESISTANCE
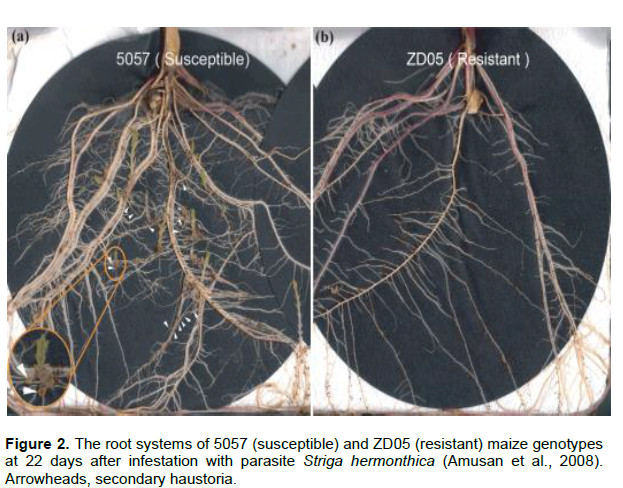
ABIOTIC STRESS RESISTANCE
CONCLUSION AND FUTURE PROSPECTS
CONFLICT OF INTERESTS
REFERENCES
|
Aditya P, Jitendra K (2014). Alien Gene Transfer in Crop Plants, Volume 2: Achievements and Impacts. Springer Science & Business Media. |
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
Amusan IO, Patrick JR, Abebe M, Thomas H, Gebisa E (2008). Resistance to Striga hermonthica in a maize inbred line derived from Zea diploperennis. New Phytol. 178:157-166. |
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
Ashraf M, Ozturk M, Ahar HR (eds) (2009). Salinity and water stress: improving crop efficiency. Springer, Berlin |
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
Bai D, Scoles GJ, Knott DR (1995). Rust resistance in Triticum cylindricum Ces. (4x, CCDD) and its transfer into durum and hexaploid wheats. Genome 38:8-16. |
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
Bennetzen JL (2007). Patterns in grass genome evolution. Curr. Opin. Plant Biol. 10:176-181. |
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
Bergquist RR (1979). Selection for disease resistance in a maize breeding programme. II. Introgression of an alien genome from Tripsacum dactyloides conditioning resistance in Zea mays. Proceedings of the tenth meeting of the Maize and Sorghum Section of Eucarpia, Varna, Bulgaria. Pp. 200-206. |
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
Bergquist RR (1981). Transfer from Tripsacum dactyloides to corn of a major gene locus conditioning resistance to Puccinia sorghi. Phytopathology 71:518-520. |
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
Berthaud J, Savidan Y, Barré M, Leblanc O (1997). Tripsacum. In. D. Fuccillo, Sears L, Stapleton P, Eds., Biodiversity in Trust. Cambridge University Press, Cambridge. Pp. 227-233. |
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
Berthaud J, Savidan Y, Leblanc O (1995). Tripsacum: diversity and Bird RMK (2000). A remarkable new teosinte from Nicaragua: growth and treatment of progeny. Maize Genetics Cooperation Newsletter 74:58-59. |
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
Branson TF (1971). Resistance in the grass tribe Maydeae to larvae of the western corn rootworm. Ann. Entom. Soc. Am. 64:861-863. |
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
Brar DS (2005). Broadening the gene pool of rice through introgression from wild species. In. Toriyama, K., Heong, K.L., Hardy, B., ed., Rice is life: Scientific perspectives for the 21st century, Proceedings of the World Rice Research Conference, Tokyo and Tsukuba, Japan, November 4–7, 2004. |
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
Canci H, Toker C (2009). Evaluation of annual wild Cicer species for drought and heat resistance under field conditions. Genet. Resour. Crop Evol. 56:1-6. |
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
Chittaranjan K (2011). Wild crop relatives: Genomic and breeding resources: Cereals. Springer Science & Business Media. |
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
Clark RB, Alberts EE, Zobel RW, Sinclair TR, Miller MS, Kemper WD, Foy CD (1996). Eastern gamagrass (Tripsacum dactyloides) root penetration and chemical properties of claypan soils. In: JE Box Jr, Ed., Root Demographics and Their Efficiencies in Sustainable Agriculture, Grasslands and Forest Ecosystems. Kluwer Acad. Pub., Dordrecht, The Netherlands. Pp. 191-211. |
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
Clifford BC (1995). Diseases, pests and disorders of oats. In: Welch RW (ed) The oat crop: production and utilization. Chapman & Hall, London, UK, Pp. 252-278. |
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
Cohen JI, Gallinat WC (1984). Potential use of alien germplasm for maize improvement. Crop Sci. 24:1011-1015. |
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
Comis D (1997). Aerenchyma: lifelines for living underwater. Agric. Res. 45:4-8. |
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
Coyne PI, Bradford JA (1985). Comparison of leaf gas exchange and water-use efficiency in two Eastern gamagrass accessions. Crop Sci. 25:65-75. |
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
De Wet JMJ (1979). Tripsacum introgression and agronomic fitness in maize (Zea mays L.). Proc. Conf. Broadening Genet. Base Crops, Pudoc, Wageningen. |
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
De Wet JMJ, Harlan JR (1972). Origin of maize: tripartite hypothesis. Euphytica 21:271-279. |
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
De Wet JMJ, Brink DE, Cohen CE (1983). Systematics of Tripsacum section Faciculata (Gramineae). Am. J. Bot. 70:1139-1146. |
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
De Wet JMJ, Harlan JR, Lambert RJ, Engle LM (1972). Introgression from Tripsacum into Zea and the Origin of Maize. Caryologia 25(1):25-31. |
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
Dela Vina AC, Mendoza ACA, Eagle LM, Ramirez DA (1995). Inheritance of selected morphological characters in Zea I. Zea mays ssp. mays × Zea mays ssp. mexicana and Zea mays ssp. mays × Zea diploperennis. Philipp. J. Crop Sci. 20:94-107. |
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
Dillon SL, Lawrence PK, Henry RJ (2005). The new use of Sorghum bicolor-derived SSR markers to evaluate genetic diversity in 17 Australian Sorghum species. Plant Genet. Res 3(1):19-28. |
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
Dillon SL, Shapter FM, Henry RJ, Cordeiro G, Izquierdo L, Lee LS (2007). Domestication to crop improvement: genetic resources for Sorghum and Saccharum (Andropogoneae). Ann. Bot. 100:975-989. |
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
Doebley JF (1990a). Molecular systematics of Zea (Gramineae). Maydica 35:143-150. |
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
Doebley JF (1990b). Molecular evidence for gene flow among Zea species. Bioscience 40:443-448. |
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
Eubanks MW (1997). Molecular analysis of crosses between Tripsacum dactyloides and Zea diploperennis (Poaceae). Theor. Appl. Genet. 94:707-712. |
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
Eubanks MW (2001). The origin of maize: evidence for Tripsacum ancestry. In. Janick J, Ed., Plant breeding reviews. John Wiley & Sons, Inc., New York 20:15-66. |
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
Eubanks MW (2002). Investigation of novel genetic resource for rootworm resistance in corn. In. NSF (ed) Proceedings of the NSF design, service and manufacturing conference. Iowa State University, San Juan, Puerto Rico, Pp. 2544-2550. |
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
Eubanks MW (2006). A genetic bridge to utilize Tripsacum germplasm in maize improvement. Maydica 51:315-327. |
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
Feldman M, Kislev ME (2007). Domestication of emmer wheat and evolution of free-threshing tetraploid wheat. Isr. J. Plant Sci. 55:207-221. |
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
Findley WR, Nault LR, Styer WE, Gordon DT (1982). Inheritance of maize chlorotic dwarf virus resistance in maize × Zea diploperennis backcrosses. Maize News Lett. 56:165-166. |
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
Gavrilova O, Gagkaeva T, Burkin A, Kononenko G, Loskutov I (2008). Susceptibility of oat germplasm to Fusarium infection and mycotoxin accumulation in grains. In. Proceedings of the 8th international oat conference, 27 June–2 July 2008, Minneapolis, MN, USA, Poster V-2a. |
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
Gill BS, Li W, Sood S, Kuraparthy V, Friebe SKJ, Zhang Z, Faris JD (2007). Genetics and genomics of wheat domestication-driven evolution. Isr. J. Plant Sci. 55:223-229. |
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
Gurney AL, Grimanelli D, Kanampiu F, Hoisington D, Scholes JD, Press MC (2003). Novel sources of resistance to Striga hermonthica in Tripsacum dactyloides, a wild relative of maize. New Phytol. 160: 557-568. |
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
Hajjar R, Hodgkin T (2007). The use of wild relatives in crop improvement : A survey of developments over the last 20 years. Euphytica 156:1-13. |
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
Hannes D, Ruth JE, Luigi G, Colin KK, Jonas VM, Jane T (2014). Adapting Agriculture to Climate Change: A Global Initiative to Collect, Conserve, and Use Crop Wild Relatives. Agroecology Sustain. Food Syst. 38(4):369-377. |
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
Harinder KC , Vineeta K , Shoukat AR (2014). Maize.. In: Aditya P, Jitendra K. Alien Gene Transfer in Crop Plants, Volume 2 Achievements and Impacts. Springer. Pp. 27-50. |
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
Hitchcock AS (1951). Manual of grasses of the United States, Second edition, revised by A. Chase. U. S. Government Printing Office, Washington, DC. |
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
Hooker AL, Perkins JL (1980). Helminthosporium leaf blights of corn the state of the art. Proceedings of the annual Corn and Sorghum Research Conference, 35:68-87. |
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
Iltis HH, Benz BF (2000). Zea nicaraguensis (Poaceae), a new teosinte from Pacific coastal Nicaragua. Novon 10:382-390. |
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
Iltis HH, Doebley JF (1980). Taxonomy of Zea (Gramineae). II. Subspecific categories in the Zea mays complex and a generic synopsis. Am. J. Bot. 67:994-1004. |
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
Iltis HH, Doebley JF, Guzman RM, Pazy B (1979). Zea diploperennis (Gramineae): a new teosinte from Mexico. Science 203:186-188. |
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
Ishimaru T, Hirabayashi H, Ida M, Takai T, San-Oh YA, Yoshinaga S, Ando I, Ogawa T, Kondo M (2010). A genetic resource for early-morning flowering trait of wild rice Oryza officinalis to mitigate high temperature-induced spikelet sterility at anthesis. Ann. Bot. 106:515-520. |
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
Kamala V, Singh SD, Bramel PJ, Manohar Rao D (2002). Sources of resistance to downy mildew in wild and weedy sorghums. Crop Sci. 42:1357-1360. |
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
Karn A, Gillman JD, Flint-Garcia SA (2017). Genetic analysis of teosinte alleles for kernel composition traits in maize. G3 (Bethesda) pii: g3.117.039529. |
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
Kemper WD, Alberts EE, Foy CD, Clark RB, Ritchie JC, Zobel RW (1997). Aerenchyma, acid tolerance, and associative N fixation enhance carbon sequestration in soil. In: R Lal, JM Kimble, RF Follett, BA Stewart., Eds., Management of Carbon Sequestration in Soil. CRC Press, Boca Raton, FL. Pp. 221-234. |
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
Kim SK, Akintunde AY, Walker P (1999). Responses of maize inbreds during development of Striga hermonthica infestation. Maydica 44:333-339. |
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
Kindiger BK, Beckett JB (1990). Cytological evidence supporting a procedure for directing and enhancing pairing between maize and Tripsacum. Genome 33:495-500. |
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
Kuhlman LC, Burson BL, Klein PE, Klein RR, Stelly D, Price HJ, Rooney WL (2008). Genetic recombination in Sorghum bicolor × S. macrospermum interspecific hybrids. Genome 51:749-756 |
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
Lagoke STO, Parkinson VO, Agunbiade RM (1991). Parasitic weeds and control methods in Africa. In: Kim SK, ed. Combating Striga in Africa, proceedings of the international workshop organized by IITA, ICRISAT, and IDRC. Ibadan, Nigeria: IITA, 3-14. |
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
Lane JA, Child DV, Moore THM, Arnold GM, Bailey JA (1997). Phenotypic characterisation of resistance in Zea diploperennis to Striga hermonthica. Maydica 42:45-51. |
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
Leblanc O, Grimanelli D, Gonzalez DLD., Savidan Y (1995). Detection of the apomixis mode of reproduction in maize Tripsacum hybrids using maize RFLP markers. Theor. Appl. Genet. 90:1198-1203. |
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
Liu Z, Cook J, Melia-Hancock S, Guill K, Bottoms C, Garcia A, Ott O, Nelson R, Recker J, Balint-Kurti P, Larsson S, Lepak N, Buckler E, Trimble L, Tracy W, McMullen MD, Flint-Garcia SA (2016a). Expanding maize genetic resources with pre-domestication alleles: Maize-teosinte introgression populations. Plant Genome 9:1.
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Copyright © 2024 Author(s) retain the copyright of this article.
This article is published under the terms of the Creative Commons Attribution License 4.0