Full Length Research Paper
ABSTRACT
Genetic relationships among 18 regenerated tall coconut accessions from International Coconut Genebank for Africa and Indian Ocean (ICG-AIO) located at Côte d’Ivoire were studied. Analyses were achieved from 17 quantitative characters of the reliable minimal list of agro-morphological descriptors for coconut proposed by Coconut Genetic Resources Network (COGENT) in 2007. From achieved Multivariate Analyses (MVA) two geographical clusters including Afro-Indian and Far East were observed in the first generation of regenerated tall coconut accessions. In addition regenerated tall coconut accessions whose parents come from South Pacific geographical area were the more varied. This typology is similar to the one of the initial introductions previously established. Thus, creation of improved hybrids from heterosis effect searching a long time exploited in tall coconut accessions can always be pursued with regenerated accessions in Côte d’Ivoire coconut program.
Key words: Tall regenerated accessions, minimum list of coconut descriptors, morphological typology, multivariate analyses (MVA), Côte d’Ivoire.
Abbreviation: COGENT: Coconut Genetic Resources Network; ICG-AIO: International Coconut Genebank for Africa and Indian Ocean; MVA: Multivariate Analyses.
INTRODUCTION
Coconut (Cocos nucifera L.), is an oleaginous, monocotyledonous plant from Arecaceae family (Teulat et al., 2000) and cultivated in wet tropical area. Coconut is used like food plant in many countries and is supplier of fatty acid to oil industry. In the Côte d’Ivoire economy, coconut palm is a very important speculation to reduce poverty at the littoral. In this area more than 20 000 families depend to coconut farming (Assa et al., 2006). In 2017, the area under coconut in Côte d’Ivoire was 103 239 ha (FAOSTAT, 2017). To sustain coconut sector, the researchers created improved coconut varieties. Coconut breeding program started by the mass selection of the best individuals from local ecotype West African Tall (WAT) (Bourdeix, 1988) that produced an average of 0.6 ton of coprah ha-1 year-1. So, annual coprah output reached 1.2 ton/ha. In the worry aim to increase the genetic gain at Côte d’Ivoire, breeders opted for the widening of the genetic basis by exotic coconut population introduction in field genebank in order to landing the inbreeding depression effect (Bourdeix, 1990). Thus, 37 accessions of the tall coconut populations and 16 populations of the dwarf were introduced from inter-tropical area between 1960 and 1980 (Fremond and De Nuce, 1971). Those coconut accessions which are conserved at field became old. The inflorescences located in foliar crown of the tall coconut palms that height exceeds 12 m become inaccessible (Bourdeix et al., 2010). Consequently, from 1987 the regeneration of aging accessions is regularly undertaken by controlled pollinated method (Konan et al., 2008) to carry out seed productions.
Today, agro-morphological diversity of the first generation cycle of regenerated accession of tall coconut palms planted in ICG-AIO located at Côte d’Ivoire is unknown. However, the knowledge of the agro-morphological traits about these regenerated accessions is important, not only for efficient management and conservation of coconut resources but also for pursue coconut breeding program in Côte d’Ivoire. Indeed, during regeneration of old coconut accession with controlled pollination method the risk of genetic diversity erosion seems to exist. Such modifications due to the artificial selection on the parental accession of plants were reported (Johnson et al., 2002; Reddy et al., 2006). In ICG-AIO, several chronological studies about agro-morphological characterization of the tall coconut accessions initially introduced were achieved (De Nuce and Wuidart, 1979; De Nuce and Wuidart, 1981; Sangaré et al., 1984; N’Cho et al., 1993; Konan, 2008) from varied quantitative descriptors (IPGRI, 1995). Basing of the multivariate analysis approaches the works of N’Cho et al. (1993) showed significant differences among seventeen tall coconut cultivars from 24 vegetative traits. Also, important morphological variability in coconut ecotypes coming from Pacific was found. Based on observations of seventeen morphological traits, Konan (2008) was classified tall coconut accessions in ICG-AIO under two groups. The first group is composed of the accessions of african origin and the Indian ocean. The second group consists of the accessions coming from South Pacific, the extreme Orient and Latin America.
However, these studies remain partial because they don't take into account the different generations of accessions in coconut field genebank. However, little information is available concerning morphological variation between and within first generation (G1) of tall coconut accessions planted in coconut genebank hosted by Marc Delorme research station in Côte d’Ivoire. Besides, during morphological diversity studies in coconut palms, the number and nature of the morphological descriptors used are varied. Considering the time that can take for the characterization of a high sample of accessions, COGENT recommended in 2007 a minimal list of 17 quantitative descriptors (www.bioversityinternational.org). Then, the reliability of this short list of agro-morphological descriptors for coconut diversity studies was proved in agro-climatic conditions of Côte d’Ivoire for survey about morphological diversity of coconut accessions hosted by Marc Delorme Research station of the Centre National de Recherche Agronomique (CNRA) (Yao et al., 2015).
The objective of the present study was to assess from this reliable short list for coconut descriptors established by the COGENT the relationships of morphological similarity among regenerated tall coconut accessions in ICG-AIO.
MATERIALS AND METHODS
Plant material
The analyses were done using the first cycle of 18 tall coconut accessions (Table 1) regenerated by controlled pollination method according to Konan et al. (2008). The regenerated WAT G1 accession used as control and planted in three experimental plots No. 043 (Latitude (5°15.725’ N), Longitude (3°50.236’ W) and Altitude (12 m)), No. 081 (Latitude (5°16.245’ N), Longitude (3°50.746’ W) and Altitude (14 m) and No. 091 (Latitude (5°16.373’ N), Longitude (3°50.758’ W) and Altitude (19 m)) was counted only one time because the different populations which compose it are similar at genetic level (Lebrun et al., 1998; Konan, 2008). All regenerated accessions were planted in ICG-AIO hosted by Marc Delorme research station of CNRA located at Côte d’Ivoire. The climate is Sudano-Guinean type and characterized by two dry seasons (January to April and August to September) and two rainy seasons (May to July and October to November). During the decade from 2000 to 2010 the annual total rainfall and sunniness were respectively 1827.9 mm and 2281.85 h onto Marc Delorme research station. Monthly averages during this same decade of the temperature and hygrometry were 26.29°C and 86.72%, respectively.
Experimental designs and assessment of the agro-morphological characters
The study was conducted on three plots for coconut genetic resources conservation. Coconut palms were planted with density of 143 trees ha-1. The experimental plots, No. 81 and No. 91, were conceived in Fisher blocks with 6 repetitions of 24 trees (4 rows of 6 trees). For these two plots, 5 healthy plants were selected randomly in each elementary plot, either a total of 30 trees per accession. The third plots No. 43 was planted in alternated rows design and the number of individuals sampled was varied from 8 to 20. In all plots, a total of 558 individuals were observed (Table 1). All observations were done from quantitative characters of the minimal list of coconut descriptors proposed by the COGENT (www.bioversityinternational.org) described in detail by Nuce and Wuidart (1982) and Yao et al. (2015). The morphological traits observed were 17 with breakdown of 4 stem, 2 floral, and 11 fruit traits.
Stem traits
Stem height (cm), bulb girth at 20 cm above soil level (cm), stem girth at 1.5 cm heigh and Height between 11 leaf scars (cm) were observed on the stem.
Floral traits
Number of female flowers and number of spikelets were counted on inflorescence of rank n.10.
Fruits traits
Fruit weight (g), husked nut weight (g), shell weight (g), endosperm thickness (mm), husk weight (g), water weight (g), endosperm weight (g), copra weight per nut (g), dry meat oil content (%), number of bunches per palm per year and number of fruits harvested per palm per year were assessed.
Data analysis
The relationships between regenerated tall coconut accessions were carried out by fours Multivariate analyses (MVA) methods (Karsai et al., 2000) using Statistica version 7.1 software (StatSoft France, 2005). Principal components analysis (PCA) was performed from the first two principal components were obtained. PCA is a data reduction method used to reduce the number of characters and to detect structure in the relationships between these characters. Cluster analysis (CA) was performed using Euclidean distance and an average fusion strategy (UPGMA) as indicators of similarity and aggregation procedure, respectively. The clusters were then represented in a dendrogram. The differences between groups established from CA were tested with Multivariate Analysis of Variance (MANOVA) at 5% threshold of likelihood. Discriminant analysis (DA) was done to determine morphological distances like Mahalanobis distances between defined clusters (Ivandro et al., 2014).
All the previously stated analyses were done on matrix of weighted mean values of each of the 17 agro-morphological characters per accession obtained from the mean value of the WAT G1 accession used as control and planted in each experimental plot. Weighting method permitted to minimize the environmental effects on agro-morphological characters expression at the accessions planted onto different experimental plots. The raw of mean data collected for each agro-morphological descriptor per accession on three distinct experimental plots were weighted from relation:
with 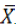 i weighted the weighted mean of the descriptor i,
i weighted the weighted mean of the descriptor i,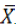 Ci the mean of the WAT G1 control on all the plots studied for the descriptor i,
Ci the mean of the WAT G1 control on all the plots studied for the descriptor i,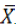 ij the raw mean of the descriptor i in the plot j and
ij the raw mean of the descriptor i in the plot j and 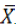 Cij the mean average of the descriptor i for the WAT G1 control in plot j.
Cij the mean average of the descriptor i for the WAT G1 control in plot j.
RESULTS
Agro-morphological variation and dendrogram
KMO (Kaiser-Meyer-Olkin) index that was superior to 0.7 indicates that the factorization of the 17 studied characters is a statistically acceptable solution. In the same way the Bartlett sphericity test on all characters showed that the factorial model is appropriated (Chi-square = 478.89, p<0,001). So, the first three principal factors that presented an eight value superior to 1 following Kaizer method accounted for 70.48% of variation were implied in variability of the regenerated accessions of tall coconut palms (Table 2).
The factor 1 expressed 38.91% of the variation. It presented a positive correlation with FW, HNW, SW, WW, EW and CWN. The component 1 can be defined as factor characterized by fruit components. The factor 2 accounted 21.08% of the total variation. It was explained by the variables, SH and L11LS that there are correlated negatively and the variable NFF that there is correlated positively. The component 2 can be defined as vegetative growth factor. The character NBPY contributed to the formation of the component 3 accounted 10.48% of the total variation (Table 2).
Dendrogram showed two morphological clusters within regenerated tall coconut accessions studied (Figure 1). The two geographical clusters, Afro-Indian and Far East, were distinguished at 600 Euclidean distances. The dendrogram showed that South Pacific accessions were met in both two clusters.
Differences between two geographical clusters “Afro-Indian” and Far East”
The difference between two majors groups according to agro-morphological characters used was significant following the multiple analysis of variance (MANOVA) at 5% likelihood (F = 8.08, p = 0.027). Data exam showed that this difference comes from 8 of the 17 characters analyzed (Table 3). These 8 characters that were essentially those describing the production such as fruit weight (FW), husked nut weight (HNW), shell weight (SW), water weight (WW), endosperm weight (EW), coprah weight per nut (CWN), dry meat oil content (DMOC) and number of fruit per palm per year (NFPY) were permitted a complete distinction of the two geographical clusters (Table 3).
Afro-Indian cluster was composed of 6 regenerated accessions that have a high yield of small fruit and oil. This group contains WAT G1, one accession coming from Indian ocean (SLT G1) and 4 accessions coming from South Pacific area (TONT G1, TAT G1, SIT G1 and VTT G1).
Far East cluster was composed of 12 regenerated accessions producing heavy fruits with high coprah content. It contains all 9 regenerated accessions coming from Far East (THT1 G1, THT4 G1, KAT4, KAT5, KAT7 G1, KAT8 G1, KAT9 G1, KAT10 G1 and RTT G1) and 3 accessions from South Pacific (RIT G1, TAGT G1 and RGT G1).
Lambda Wilks’test from discriminant analysis, associated to the hypothesis of equality of the middle vectors between two agro-morphological groups was significant at 5% likelihood (F = 0.234, p = 0.039). So the two major groups established from cluster analysis were different. Mahalanobis distance between two agro-morphological clusters (182.81 Mahalanobis units) was significant at 5%
likelihood (F = 9.234, p = 0.043).
DISCUSSION
The observed differences between coconut populations with regard to agro-morphological traits are indirect or direct representations of differences at the DNA level. For this study, agro-morphological clustering of the first generation G1 of regenerated accessions of tall coconut palms was analyzed. The investigations revealed two agro-morphological clusters into these tall accessions that are Afro-Indian and Far East. This clustering could reveal two biological realities.
Firstly, the studied morphological descriptors seem to discriminate tall coconut populations with the same level like foliar polyphenols markers (Jay et al., 1989) and the genetic markers such as RFLP (Lebrun et al., 1998) and SSR (Konan, 2008). These genetic markers were permitted in some cases to group in same way coconut populations hosted by international genebank at Côte d’Ivoire. These studies oppose an Afro-Indian cluster to another compound of the accessions that come from South Pacific, Far East and Latin America. The reliability of this tall coconut populations clustering would be due to biological data provide to fruit component descriptors and the weighting method in relation to fragmented means of WAT G1 control used. Indeed, the study of N’cho et al. (1993) achieved on the same origins of coconut populations used in absence of the fruit component descriptors and no correction of raw data by weighting method was generated less clean of tall coconut clustering in nine morphotypes. Using of the weighing method of raw mean data of each descriptor assessed on tall coconut accessions planted onto different plots in relation to fragmented means of WAT G1 control is minimized environment effects on characters expression (Bourdeix, 1989). Thus, the expression of the phenotypes due to the main effects of the genes has been only revealed.
Secondly, the morphological clustering of the first cycle (G1) of tall coconut accessions regenerated by controlled pollination method with the one of the initial introductions (G0) was conform (Konan, 2008). Indeed, the sexual reproduction involves by control pollinations between male and female individuals chosen as parents would protect the organisms against eventual mistakes of DNA replication (Guetet et al., 2008). That conferred to the regenerated accessions (G1) an adaptation to climatic change and modifications concerning soil chemical properties. According to Ivandro et al. (2014), genetic distance measures based on phenotypic characters are one of the main multivariate techniques used to provide criteria for choosing parents. Thus, the parental tall coconut accessions to choose in the crossings should belong to the two revealed groups. There is a high probability of selecting transgressive genotypes due to the occurrence of heterosis and the action of complementary dominant genes (Ivandro et al., 2014). Also, the morphological structure of the regenerated accessions was revealed that those whose parents come from South Pacific are the more varied as one reported by N’cho et al. (1993) and Sugimura et al. (1997) in coconut genebank of Côte d’Ivoire and Philippines respectively.
CONCLUSION
This survey was initiated to evaluate regenerated tall coconut diversity using reliable short list of agro-morphological descriptors. The results revealed two genetically pools into regenerated Tall coconut accessions in ICG-AO, Afro-Indian and Far East. Also, it appears from analysis of the present results in relation to the findings of the previous research works that the agro-morphological clustering of the regenerated Tall coconut accessions (G1) was conform to the one of the parental accessions (G0) previously established in ICG-AO. Until further DNA study at large scale including all regenerated Tall coconut accessions planted in ICG-AO, if the morphological distances between these two groups of regenerated accessions reflect in fact the genetic distances, creation of improved coconut hybrids from heterosis effect searching a long time exploited in Tall coconut accessions can be pursued in coconut breeding program of Côte d’Ivoire
CONFLICT OF INTERESTS
The authors have not declared any conflict of interests.
REFERENCES
|
Assa R. Konan K, Nemlin J, Prades A, Agbo N. Sie R (2006). Diagnostic de la cocoteraie paysanne du littoral ivoirien. Sciences et Nature. Série A, Biosciences Agronomie Environnement Biotechnologie 3(2):113-120. |
|
|
Bourdeix R (1988). Efficacité de la sélection massale sur les composantes du rendement chez le cocotier. Oléagineux 43(7):283-290. |
|
|
Bourdeix R (1989). La sélection du cocotier (Cocos nucifera L.). Etude théorique et pratique. Optimisation des stratégies d'une amélioration génétique. Thèse de Doctorat, Université de Paris Sud, Centre d'Orsay (France), 194 p. |
|
|
Bourdeix R, Konan J, Saraka D, Allou K (2010). Regeneration of old coconut accessions in the International Genebank for Africa and the Indian Ocean. In: Rival Alain (ed.). Palms 2010, Biology of the palm family (Abstracts books): International Symposium 5-7 May 2010, Montpellier, France P 55. |
|
|
De Nuce L, Wuidart W (1979). Les cocotiers grands de port Bouët (Côte d'Ivoire). 1- Grand Ouest Africain, Grand de Mozambique, Grand de Polynésie, Grand de Malaisie. Oléagineux 34(7):339-349. |
|
|
De Nuce L, Wuidart W (1981). Les cocotiers Grands à Port-Bouët (Côte d'Ivoire). 2- Grand Rennell, Grand Salomon, Grand Thaïlande, Grand Nouvelles-Hébrides. Oléagineux 36(7):353-362. |
|
|
De Nuce L, Wuidart W (1982). L'observation des caractéristiques de développement végétatif, de floraison et de production chez le cocotier. Oléagineux 37(6):291-297. |
|
|
FAOSTAT (2017). [January, 31th 2019]. |
|
|
Fremond Y, De Nuce L (1971). Le bloc d'amélioration du cocotier de Port Bouët. Oléagineux 2(1):71-82. |
|
|
Guetet A, Boussaid M, Neffati M (2008). Etude de la vigueur reproductive de populations naturelles d'Allium roseum en Tunisie. Plant Genetic Resources Newsletter 153:28-35. |
|
|
IPGRI (1995). Descriptors for coconut (Cocos nucifera L.). International Plant Genetic Resources Institute (IPGRI), Rome (Italy) P 61. |
|
|
Ivandro B, Fernando I, Antonio C (2014). Parental Selection Strategies in Plant Breeding Programs. Journal of Crop Science and Biotechnology 10(4):211-222. |
|
|
Jay M, Bourdeix R, Potier F, Sanlaville C (1989). Premiers résultats de l'étude des polyphénols foliaires du cocotier. Oléagineux 44:151-161. |
|
|
Johnson R, Bradley L, Evans A (2002). Effective population size during grass germplasm seed regeneration. Crop Science 42:286-290. |
|
|
Karsai I, Meszaros K, Lang L, Hayes P, Bedo Z (2000). Multivariate analysis of trait determining adaptation in cultivated barley. Plant Breeding 120:21-22 |
|
|
Konan J, Bourdeix R, George M (2008). Directives pour la régénération: cocotier. In: Dulloo M, Thormann I, Jorge M, Hanson J editors. Crop specific regeneration guidelines [CD-ROM]. System-wide Genetic Resource Programme, Rome, Italy. |
|
|
Konan J (2008). Evaluation de la diversité agromorphologique et moléculaire de la collection internationale de cocotier (Cocos nucifera L.) en Côte d'Ivoire. PhD thesis, University of Cocody, Côte d'Ivoire, 150 p. |
|
|
Lebrun P, N'cho Y, Seguin M, Grivet L, Baudouin L (1998). Genetic diversity in coconut (Cocos nucifera L.) revealed by restriction fragment length polymorphism (RFLP) markers. Euphytica 101:103-108. |
|
|
N'cho Y, Sangaré A, Bourdeix R, Bonnot F, Baudouin L (1993). Evaluation de quelques écotypes de cocotier par une approche biométrique. 1. Etude des populations de grands. Oléagineux 48(3):121-132. |
|
|
Reddy K, Upadhyaya H, Reddy L, Gowda C (2006). Evaluation of Pollination Control Methods for Pigeonpea (Cajanus cajan (L.) Millsp.) Germplasm Regeneration. SAT eJournal, ejournal.icrisat.org 2(1). |
|
|
Sangaré A, Le Saint J, De Nuce L (1984). Les cocotiers Grands à Port-Bouet (Côte d'Ivoire). 3- Grand Cambodge, Grand Tonga, Grand Rotuma. Oléagineux 39(4):205-213. |
|
|
Sugimura Y, Itano M, Salud C, Otsuji K, Yamaguchi H (1997). Biometric analysis on diversity of coconut palm: cultivar classification by botanical and agronomical traits. Euphytica 98:29-35. |
|
|
Teulat B, Aldam C, Trehim P, Lebrun P, Barker J, Arnold G, Karp A, Baudouin L, Rognon F (2000). An analysis of genetic diversity in coconut (Cocos nucifera L.) population from across the geographical range using sequence tagged microsatellites (SSRs) and (AFLPs). Theoretical applied Genetic 100:764-771. |
|
|
Yao S, Konan J, Sie R, Diarrassouba N, Lekadou T, Koffi E, Yoboue K, Bourdeix R, Issali A, Doh F, Allou K, Zoro Bi I (2015). Fiabilité d'une liste minimale de descripteurs agromorphologiques recommandée par le COGENT dans l'étude de la diversité génétique du cocotier (Cocos nucifera L.). Journal of Animal and Plant Sciences 26(1):4006-4022. |
|
Copyright © 2024 Author(s) retain the copyright of this article.
This article is published under the terms of the Creative Commons Attribution License 4.0