ABSTRACT
Biofortification is a process in which plant species are improved in order to raise the nutritional content of the product consumed. The following are some of the main techniques that can be introduced in plant breeding programs in order to improve the nutritional quality of the food to be produced. The rice breeding for higher Fe and Zn content in grain, rice breeding for higher β-carotene content in the grain, rice breeding for higher folate content in the grain, techniques used for plant transformation, transformation via protoplasts, transformation via bioballistics (or biolistic), transformation via Agrobacterium tumefaciens, identification and in vitro selection of transformed tissues, concepts of gene expression and regulation, protein expression, proteomics, mutant study, patch-clamp, were studied. It is necessary to verify the occurrence of variability among the rice genotypes for the Fe, Zn and B complex vitamins; therefore, it will be possible to select these materials within breeding programs. The simple selection of these superior genotypes in relation to nutritional quality, even though by means of conventional breeding techniques, can bring benefits to rice consumption by the human population.
Key words: Cultivated rice (Oryza sativa), agriculture, plant breeding, genetics, transformation, selection, gene expression, micronutrient.
Estimates point to an accelerated growth of the world population reaching about 9 to 10 billion people (Godfray et al., 2010; FAO, 2017). In this context, food production should accompany such demand and is considered a fundamental factor for food security in the planet. However, micronutrient deficiency in humans is mainly caused by the deficient daily diet, combined with the low concentrations and low availability of these elements in the products consumed. Currently, deficiencies caused by the lack of iron (Fe), iodine (I), selenium (Se), vitamin A and the zinc (Zn) are the ones that cause greater concern with regard to human health. Research indicates that this deficiency affects infants, preschool children, teenagers, pregnant and women in fertile age (Zancul, 2004; Andersson et al., 2017). Among the strategies to reduce malnutrition in the world, food biofortification has been pointed out as one of the options of greater practical feasibility (White and Broadley, 2005; Sharma et al., 2017). In this context, significant progress has been achieved at International Rice Research Institute (IRRI) by enriching rice grain with provitamin A (b-carotene) and biofortification of iron and zinc (Andersson et al., 2017). Thus, bioavailability of these micronutrients will provide benefit to poor rice consumers, particularly in Asia to overcome the problems of vitamin A, iron and zinc deficiency in their diet (Zancul, 2004; Jena and Nissila, 2017).
Biofortification is a process in which plant species are improved in order to raise the nutritional content of the product consumed (grains, fruits, roots, tubers and leaves). This mechanism of plant breeding is the promising and relatively low cost (Poletti et al., 2004; Sharma et al., 2017; Bouis and Saltzman, 2017). Since agriculture is the main supplier of these micronutrients, it is essential to recognize the importance of soil and plant nutrition as influential to human health. Biofortification, through the development and production of bioavailable micronutrient enriched plants, can be a practical agronomic tool to combat nutrition problems globally (Bouis and Saltzman, 2017). In this context, the adequate crop fertilization and nutrition, mainly regarding the use of nitrogen, besides increasing the grain yield, results in the improvement of the nutritional and technological quality, inclusive on rice grain protein content (Mingotte et al., 2012, 2013, 2015). The introduction of biofortified agricultural products and improved varieties with higher mineral and vitamin content, in addition to complementing existing nutrition interventions, can provide greater sustainability and low cost for producers and consumers. The International Center for Tropical Agriculture (CIAT) and the International Food Policy Research Institute (IFPRI) coordinate the "Harvest Plus" food biofortification program, a global alliance of research institutions and executing agencies that have teamed up to improve and disseminate products of better nutritional quality.
According to Andersson et al. (2017), the HarvestPlus works in partnership with more than 200 scientific and implementation organizations around the world to improve nutrition and public health by developing and promoting biofortified food crops that are rich in vitamins and minerals, and providing global leadership on biofortification evidence and technology. The initial phase of the program includes six basic crops for human consumption: beans (Phaseolus vulgaris), cassava (Manihot esculenta), maize (Zea mays, L.), rice (Oryza sativa), sweet potatoes (Ipomoea batatas) and wheat (Triticum spp.). The objective is to generate technologies and knowledge for the development of conventional cultivars with better protein quality and higher levels of iron, zinc and provitamin A in the grains, which should be cultivated and consumed in several developing countries. The increase in bioavailability of essential elements in food or the implantation of species grown under selected soils with high concentration of these nutrients may be attractive tools in the production of foods based on biofortified plant, especially in the case of heavy metals. In the particular case of biofortification in Se, it is necessary to determine the specific forms in which this element is incorporated and metabolized by the plant, as this determines its bioavailability for human absorption and its biological activity in humans (Finley, 2006), for example, the production of selenoglutathione peroxidase, a potent antioxidant enzyme (Whanger, 2002).
The biofortified rice with minerals and provitamin A can complement nutrition, as well as being a sustainable and low-cost form of intervention, especially the economically disadvantaged populations. Studies suggest that doubling iron content in rice grain may increase the supply of this nutrient by 50% to the poor populations. Even with genotypic variations, in the grain rice, duplication of iron and zinc contents occurs (Gregório et al., 2000). The combination of high iron and zinc content in golden rice can help in the fight against malnutrition (Khalekuzzaman et al., 2006). The rice biofortification, with high levels of β-carotene, iron, zinc and lysine, combined with the development of superior agronomic and yield traits, can support the healthy eating and subsistence of millions of people who need it (Datta et al., 2006). The first step in obtaining golden rice was to isolate the three genes responsible for producing the enzymes responsible for β-carotene synthesis. Two of these genes were isolated from the daffodil (Narcissus pseudonarcissus) and a gene from the bacterium Erwinia uredovora. The isolation of these genes was carried out with restriction enzymes, that is, enzymes that act on DNA fragmentation in sequences of specific nitrogenous bases. The transformation method adopted was via Agrobacterium, in contact with embryo cells in rice seeds. The three incorporated genes pass to code for enzymes responsible for the transformation of geranyl-geranyl diphosphate into β-carotene, which makes the rice grain yellow.
In this context, Burkhardt et al. (1997) verified that the Japonica rice model variety Taipei 309 was transformed by microprojectile bombardment with a cDNA coding for phytoene synthase from daffodil (Narcissus pseudonarcissus) under the control of either a constitutive or an endosperm-specific promoter. In transgenic rice plants, the daffodil enzyme is active, as measured by the in vivo accumulation of phytoene in rice endosperm. Thus, it is demonstrated for the first time, that it is in principle, possible to engineer a critical step in provitamin A biosynthesis in a non‑photosynthetic, carotenoid‑lacking plant tissue. These results have important implications for long-term prospects of overcoming worldwide vitamin A deficiency. The following are some of the main techniques that can be inserted into the rice breeding programs in order to improve the nutritional quality of the food to be produced.
RICE BREEDING FOR HIGHER FE AND ZN CONTENT IN THE GRAIN
Approximately, 150 million hectares of rice are grown in the world, where 75% comes from the irrigated farming system. Under flooded conditions, several changes occur in soil chemical reactions, for example, the reduction of iron, which previously precipitated as hydroxides and oxides, passes to its more soluble form (Fe2+), which also increases its availability to the plants (Ponnanperuma, 1972). The concentration of soluble iron may be so high that, depending on the genotype, it can cause toxicity to the plants, leading to losses in productivity. The importance of breeding programs in the selection of rice genotypes that are tolerant to the high iron content in the soil, especially under flood conditions, is important. In the Philippines, the breeding program of the IRRI develops studies on biofortification, producing rice-enriched seeds with iron (Fe). In the case of Fe, the bioavailability of ferritin from transgenic rice has been already tested in deficient rats, and rice diets were as effective as the FeSO4 diet in hematocrit replacement (Murray-Kolb et al., 2000). Since ferritin is used as a natural source of iron in the early development of animals and plants, it has high bioavailability. Researchers have reported genetic variability in the rice for tolerance to excess Fe in the soil. Researchers concluded that the cultivar "CK4" rice tolerated conditions of excess Fe when cultivated under flooded conditions. This tolerance is due in part to the lower accumulation of Fe in the leaves and due to the superior photosynthetic potential in the presence of Fe in the foliar tissue.
Differences between the rice cultivars under Zn deficiency conditions, especially under high pH soil conditions, were associated with susceptibility to HCO3 (Forno et al., 1975). Bicarbonate concentrations of 5 to 10 mM may inhibit root growth of the inefficient rice cultivars in Zn uptake, but on the other hand, this same condition can stimulate root growth in efficient plants (Yang et al., 1994). The higher zinc acquisition by the rice plant is related to tolerance to the high concentration of HCO3 in the soil (Yang et al., 1994). The differential susceptibility between beans and soybean (Glycine max) for zinc deficiency was associated with the restricted translocation of this nutrient from the roots. Genotypic differences for zinc use efficiency have been related to the absorption efficiency and the translocation capacity of this nutrient in the roots. Under Zn deficiency, there is an increase in the release of phytosiderophores in the form of root exudates in grass species, possibly as an adaptive response to Zn deficiency. This physiological response triggers the mobilization of zinc and, consequently, a greater accumulation of this nutrient in the seeds. Impa et al. (2013) studying the internal Zn allocation influences on Zn deficiency tolerance and grain Zn loading in rice (Oryza sativa) suggested that some Zn-efficient rice genotypes have greater ability to translocate Zn from older to actively growing tissues than genotypes sensitive to Zn deficiency. In this way, quantitative trait loci (QTLs) associated with Zn enhancement in rice has been reported, but none of them have an effect larger than 30% phenotypic variation (Stangoulis et al., 2007; Norton et al., 2014). Research efforts continue to identify major QTLs associated with grain zinc content and for better understanding of zinc uptake, transport and remobilization in the grain (Palmgren et al., 2008; Andersson et al., 2017).
In relation to the limitation of the availability of Fe in the soil, the plants evolved two strategies, known as “Strategy I” and “Strategy II”, of the Fe absorption. The mechanism known as Strategy I includes extrusion of protons to solubilize Fe3+ in the solution of the soil, Fe3+ reduction solubilized by Fe3+ membrane-bound reductase chelate, resulting in the subsequent Fe2+ transport by the plant root cells by Fe2+ transporters. Several studies have shown that plants belonging to Strategy I (non-grasses) when under conditions of Fe deficiency develop morphological and physiological responses. Among the observed responses is the development of root hair, through a transition or transfer cells, increasing the contact surface with the middle that surrounds the roots. This morphological modification favors the release of protons in greater quantity by the ATPases, which are "triggered" by response genes involved in the Fe transport, acidifying the middle to make the Fe more soluble. Also, expression of FRO genes involved in the Fe-reductase activity and IRT genes encoding proteins for the Fe transport occurs (García et al., 2011). In grasses, Strategy II occurs, characterized by the formation of Fe chelates, involving the release of phytosiderophores and subsequent absorption of the Fe3+ through membrane sites specialized in their transport (Marschner, 1995). Zuo and Zhang (2011) highlighted the great genetic variability in relation to Fe content in the edible portions of most cultivated species, including wheat, beans, rice, corn, cassava, yams, medicinal herbs and lentils (Lens culinaris). This genetic variation can boost the development of plants with high Fe content for the benefit of human health. Generally, grains of cereal such as wheat and rice have lower Fe concentrations when compared with legume seeds (Frossard et al., 2000). The potential for increasing Fe content in rice grains is about four to five times, depending on the variability. When the difference between traditional and modern rice varieties is taken into account, the traditional ones contain higher Fe in the grains, while the modern varieties have lower Fe concentrations (Gregorio et al., 2000, 2008). This particular observation can be attributed to the fact that direct selection for higher Fe content in grains was not part of previous rice breeding programs (Zuo and Zhang, 2011).
RICE BREEDING FOR HIGHER β-CAROTENE CONTENT IN THE GRAIN
Vitamin A is also used in genetic engineering in the production of fortified seeds. Researchers at the University of Freiburg, Germany, who studied biofortification, introduced β-carotene into the rice endosperm to produce the "Golden Rice". Studies are being carried out to ensure that the rice enriched with β-carotene is used in developing countries to combat vitamin A deficiency (Beyer et al., 2002; Datta et al., 2007). Brown rice is rich in micronutrients and carotenoids (Tan et al., 2004), but the polishing process considerably reduces its nutritional value. The control of gene expression of ferritin on the control of the glutelin promoter in rice has been efficient in increasing nutritional levels not only in whole grains of rice but also in polished grain (Goto et al., 1999; Vasconcelos et al., 2003; Khalekuzzaman et al., 2006). Similar principles have been used in the development of the golden rice (Datta et al., 2007). Currently, none of the rice genotypes presents β-carotene contents in the polished grains. There is no doubt that there is potential in the use of genetic variability of the carotenoid content in rice grains (Tan et al., 2004). More than half of all women and children in Southeast Asia and Southern countries are anemic. Anemia limits growth and cognitive development in children and increases the incidence of death in severely anemic women during childbirth. However, the bioavailability of β-carotene should be studied with greater depth. New studies suggest that a small supplement of vitamin A or β-carotene may reach twice the endogenous absorption of iron from cereals (Graham and Rosser, 2000). Thus, consumption of β-carotene enriched rice may reduce deficiencies in vitamin A and protect against iron deficiency anemia.
RICE BREEDING FOR HIGHER FOLATE CONTENT IN THE GRAIN
Folate deficiency is a health problem that affects many people in the world and is associated with deficiency in the B-complex vitamins. Food fortification by industrial supplementation and consumption of folic acid tablets are alternatives, but may not be feasible in less developed countries. Recent advances show that biofortification of agricultural products aimed at raising folate content is a viable strategy to combat folic acid deficiency around the world. The genes and enzymes involved in folate biosynthesis are sufficiently understood. Thus, metabolic engineering mechanisms and the results of preliminary studies of genetic engineering in plants are encouraging (Bekaert et al., 2008). The steps of folate synthesis are the same in plants and bacteria, and pathway enzymes and their genes are all known in both groups (Storozhenko et al., 2007). Essentially, the three fractions of the tetrahydrofolate (THF) molecule are pteridine, p-aminobenzoate (P-ABA) and portions of glutamate. In bacteria, the process occurs in the cytosol, but in plants, three cellular compartments are involved: the plastids, mitochondria and cytosol (Jabrin et al., 2003). The plants have developed mechanisms to deal with the instability of folic acid, and it is possible to reduce its rate of degradation. In this aspect, the initial studies on tomato fruits and Arabidopsis thaliana by the expression of the enzyme cyclohidrolase I, known as GTPCH I, are promising (Diaz de La Garza et al., 2004). In both cases, with the insertion of the GenBank genes BE136861 and AE000304, the levels of pteridine in tomato (Solanum lycopersicum) and Arabidopsis (transgenic) were, respectively, 140 and 1250 times higher than the wild type, however, the increase in the content of folate was only two to four times, indicating the need for additional engineering of this metabolic pathway. In transgenic tomato plants, p-ABA reduction occurred, and with the exogenous delivery for GTPCHI overexpression, resulted in an increase in folate content, with an additional 2.5 to 10-fold (Diaz de La Garza et al., 2004). This observation points not only to the need for simultaneous reinforcement of both folate precursors (pterine and p-ABA) but also demonstrates a great physiological potential to increase folate concentration within plant cells.
TECHNIQUES USED FOR PLANT TRANSFORMATION
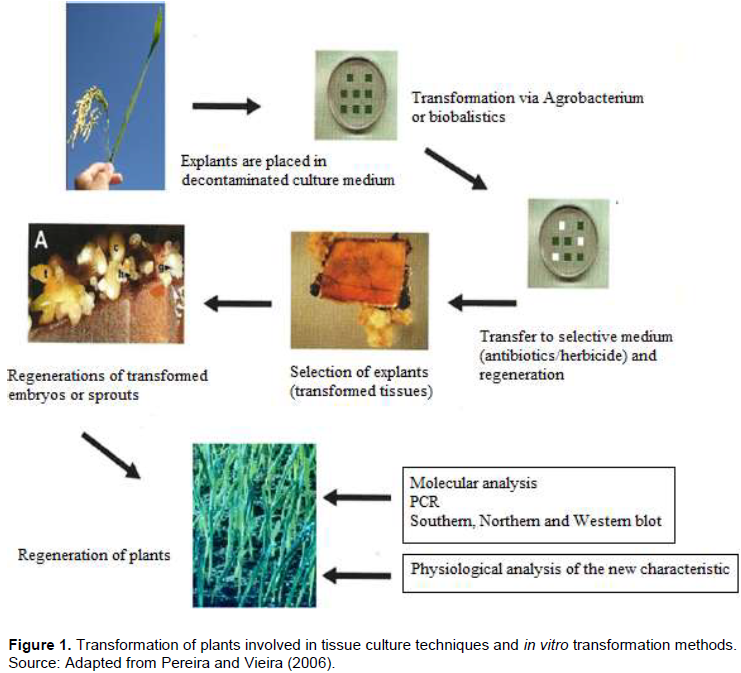
Genetic transformation of plants is one of the major advances in agricultural technology in recent years. The production of genetically modified plants (GMOs) has a great economic importance due to the possibility of incorporating new agronomic traits into the plants cultivated, such as resistance to herbicides, insects or phytopathogens, as well as improvement in food quality (Pereira and Vieira, 2006). The transformation of plants combines techniques of tissue culture, methods of transformation and selection in vitro (Figure 1). The transformation can be done directly by the physical introduction of DNA into plant cells, or by biological methods, where microorganisms such as viruses or bacteria are used as transformation vehicles. Three transformation systems are more widely known and therefore common, these are, transformation via protoplasts, bioballistics and via Agrobacterium tumefaciens. Nowadays different characteristics of socio-economic interest have already been introduced in different plant species by genetic transformation. These characteristics mainly aimed at improving performance in the field of cultivated plants, by means of resistance to biotic and abiotic stresses. Characteristics related to the development of the plant and the quality of the product can also be modified in transgenic plants. The trend is to increase the number of traits that can be manipulated through genetic engineering, increasing the range of products to be made available to the farmer and the consumer.
Transformation via protoplasts
Protoplasts, plant cells without a cell wall, can be considered as the ideal type of plant cell for transformation. The absence of the cell wall allows free transit to a DNA sequence that can penetrate most cells, including the competent cells for transformation and regeneration (Potrykus, 1991). This method of transformation can be carried out chemically, using polyethylene glycol or polyvinyl alcohol, or by electroporation, which consists of producing pores in the membrane through fast high voltage electrical impulses. As a limitation of the transformation technique via protoplasts, it is worth mentioning, the difficulty in regeneration, because few plant species have a high regenerative rate. The major success of protoplast transformation was observed in rice, where transgenic plants with hygromycin phosphotransferase and b-glucuronidase (GUS) genes were obtained for both indica and japonica subspecies (Zhang and Wu, 1988).
Transformation via bioballistics
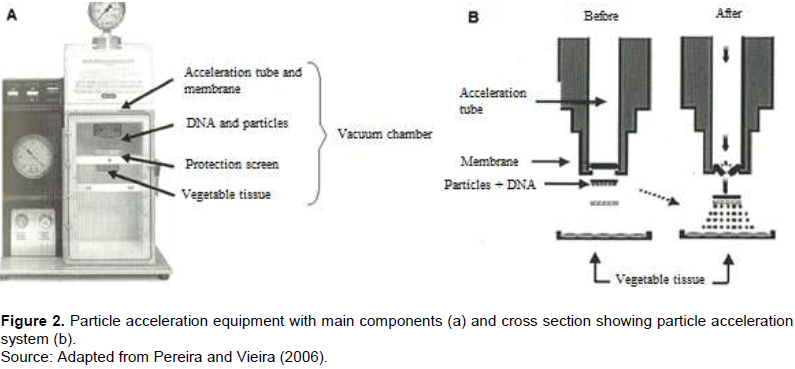
The insensitivity of certain plant species, especially monocotyledons and angiosperms, to Agrobacterium infection limits the use of this bacterium as a vector of genetic transformation. Thus, direct transformation systems, based on chemical or physical methods, were developed in parallel with the Agrobacterium system. The bioballistics (or biolistic) method, or particle bombardment, has been highlighted as one of the most important methods of transformation of plants because, in addition to transforming plant tissues, it also allows the transformation of microorganisms or animal tissues (Klein et al., 1992). It is a technique with potential for use in any plant species, provided there is an in vitro regeneration system from the target tissue. Many devices have been proposed for particle acceleration. Nowadays, the most used models are the acceleration of particles by electric discharge or air pressure, using membranes of different thickness (Figure 2). In comparison of this method with the Agrobacterium transformation system, bioballistics does not depend so much on the genotypes and their interaction with the bacteria, but rather, there is a good in vitro regeneration system of the material to be transformed (Pereira and Vieira, 2006).
Transformation via A. tumefaciens
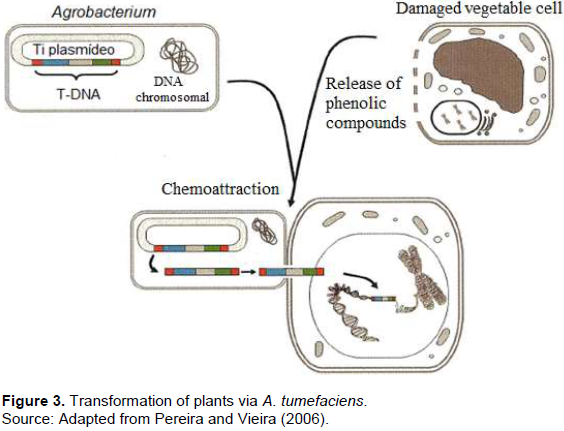
The transformation methods mostly used in the production of GMO plants are mediated by the A. tumefaciens and via bioballistics. The Agrobacterium is a typical soil bacterium that has the ability to transfer DNA fragments into plants, and it has been used as a plant transformation vector. Obtaining a transgenic plant involves the transfer and integration of the transfer DNA (T-DNA) into the plant cell and the ability of these transformed cells to differentiate into a plant (Figure 3). The ability of differentiation, called totipotency, allows the regeneration of plants by in vitro tissue culture techniques. The advancement in molecular biology knowledge is fundamental both to elucidate the in depth molecular bases of the Agrobacterium-host interaction process and to build transformation vectors based on the tumor-inducing (Ti) plasmid. Thus, molecular biology techniques in association with in vitro plant tissue culture techniques form the basis for obtaining a transgenic plant.
Identification and in vitro selection of transformed tissues
In studies on genetic transformation of plants, the use of reporter genes or marker genes is necessary, besides the genes of agronomic or scientific interest. Reporter genes serve to verify that the transformation process is being done correctly by identifying transformed cells or tissues. These genes allow an analysis of transient expression of the gene temporarily, even when the gene is not integrated into the genome of the cells. The marker genes are used to enable discrimination between transformed and untransformed cells, and consequently the selection of the former. Such genes are introduced to facilitate the work of identifying them because they are a minority in relation to the total cells undergoing a transformation. Antibiotic resistance genes are generally used. Thus, at the time of plant regeneration from a cell, the addition of antibiotic to the middle will only allow the growth of the transformed cells expressing the right protein. One of the most commonly used reporter genes is uidA which encodes the b-glucuronidase enzyme (GUS) and
provides a histochemical or fluorescent analysis of the transformed material. Other widely used reporter genes are green fluorescent-protein genes (gfp), extracted from Jellyfish Aequorea victoria; and the Luciferase gene (luc), isolated from Photinus pyralis (firefly). These reporter genes (luc and gfp) have been most widely used in the identification of transformed tissues. With the development of more sensitive photodetection and imaging equipment, luc genes are being used for in vivo labeling and visualization in real time of cells and tissues; for marking and dissemination studies of pathogenic microorganisms in plants and animals; as sensitive markers of cancer cells, assisting in the study of metastatization and the development of new therapies; and as environmental biosensors for the detection of heavy metals such as mercury and arsenic or pesticides in contaminated waters (Viviani, 2008). One of the most recent applications in the field of proteomics involves the use of luc and gfp, because the methods that involve reporter genes allow the identification of the site (root, leaf or fruits) of occurrence of gene expression, being related to gene regulation. Methods related to the expression allow evaluating the occurrence of gene expression in certain situations, such as the presence or absence of a certain nutrient, which is possible for selecting genotypes.
CONCEPTS OF GENE EXPRESSION AND REGULATION
The analysis of gene expression is importance in many fields of biological research. Knowledge on gene expression patterns should provide a better understanding of complex regulatory systems and probably lead to the identification of relevant genes for plant breeding. The discovery of the polymerase chain reaction (PCR) has brought enormous benefits and scientific development, such as genome sequencing and gene expression. This technique allows amplifying a specific segment of a DNA molecule using two primers (short nucleotide sequences) complementary to the ends of the segment to be amplified and a DNA polymerase. The technique called RT-PCR allows amplifying the coding passages directly from mRNA molecules. After extraction of the RNA, a single strand of DNA complementary to the mRNA (cDNA) is synthesized using the reverse transcriptase enzyme of viral origin. The RNA tape from The RNA / DNA hybrid is digested with RNase, and the cDNA strand is replicated by a DNA polymerase. When compared with the other two techniques commonly used to quantify mRNA, Northern blot analysis and RNase protection assay, RT-PCR can be used for the quantification of mRNA in much smaller samples. In fact, this technique is sensitive enough to allow approximate quantification of RNA from a single cell (Weis et al., 1992).
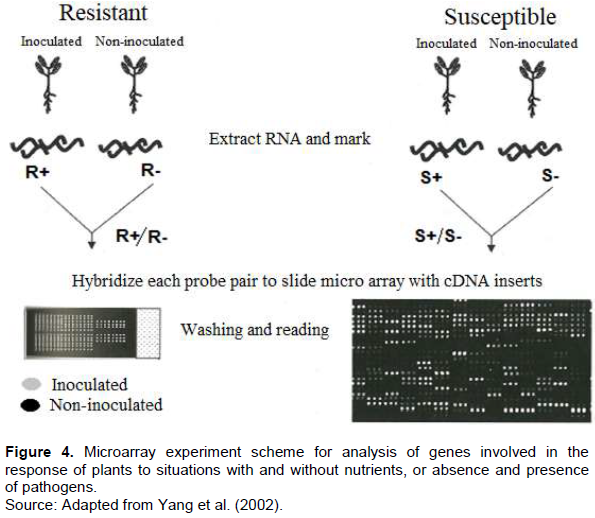
The technological innovation resulting from PCR, called real-time PCR, has been gaining ground in clinical diagnostics and research laboratories because it has the ability to obtain results more accurately and rapidly in relation to PCR. Several methodologies have been developed for the large-scale identification of differentially expressed genes. Among the most used, the microarray technique, which is based on the quantification of mRNA produced by the cell (gene expression products), provides a powerful platform for analyzing the expression pattern of thousands of genes simultaneously. In the microarray technique, the mRNA is extracted from the tissues to be studied comparatively. The mRNAs are transformed into cDNA. Each cDNA pool is marked with clusters of fluorescent nature. After labeling, the samples in the microarrays are denatured by heat. The cDNAs studied are grouped and placed in contact with the microarrays by submerging the chip in a hybridization solution. Both cDNAs of the tissues studied and the "pores" of the slide are in plain ribbon form. If there is complementarity between the sequences, the labeled cDNA strands will hybridize to the cDNAs of a "pore" of the slide. After washing and drying the chip, it is taken to a dark room and irradiated with a laser. The fluorescent markers will absorb the radiation. Each marker emits radiation at a different wavelength, allowing the quantification of hybridized cDNA in any "pore" of the slide.
As gene fixed in each of the "pores" is known, it is possible to know the expression of a control tissue in relation to the variant. Colors are assigned computationally to the emission ranges of each of the markers and the variations between the colors determine the levels of expression (Figure 4) (Yang et al., 2002). The microarray technique allows the analysis, in parallel, of thousands of genes in two populations of labeled RNAs, while RT-PCR provides the simultaneous measurement of gene expression in different samples for a limited number of genes and it is especially desirable when only a small number of cells are available. Both techniques have the advantage of speed and the high degree of potential automation when compared with traditional quantification methods such as Northern blot analysis, in situ hybridization and ribonuclease protection assay. In situ hybridization is a method of locating and detecting specific nucleic acid sequences (DNA and RNA) of tissue sections preserved or prepared from cells by hybridizing a complementary band of a specific nucleotide probe (DNA or RNA) of the sequence of interest. This technique, while being a complex method, allows a localization of specific cell transcripts within a tissue. The basic principles for in situ hybridization are the same for DNA or RNA, varying in type of cells and tissues to be studied, which require probes to detect specific nucleotide sequences within cells and tissues (Parker and Barnes, 1999).
Protein expression
The identification of specific proteins has been one of the main objectives of scientific and diagnostic practices. Blotting techniques are used to identify unique proteins or sequences of nucleic acids. The three main blotting techniques that have been developed are commonly called Southern, Northern and Western Blotting, and they enable the identification of DNA, RNA, and proteins, respectively. Southern blotting (Southern, 1975) allows DNA fragments to be identified with DNA probes, which are hybridize by hydrogen bonding to form complementary fragments of chromosomal DNA. Prior to Southern blotting analysis, DNA fragments must be produced from chromosomes by enzymatic digestion with restriction endonucleases. These enzymes are obtained from microorganisms and digest the double-stranded DNA at specific sites determined by the nucleic acid sequence. Northern blotting analysis employs essentially the same procedure described for Southern blotting, except that complementary DNA is used for RNA probe. Northern blotting technique (Thomas, 1983) allows individual messenger RNA molecules (mRNA) be identified and measured after hybridization of their corresponding DNA sequences. The messenger RNA is initially separated by electrophoresis based on its size and they are transferred to paper before the probes genes that are used to locate the RNA of interest. Western blotting allows specific proteins to be identified with specific antibodies used as analytical probes. The Western blotting analysis requires specific antibodies for the protein under analysis.
Proteomics
Although, the identification of all proteins encoded in the genome of an organism seems a rather difficult task to perform, even in simpler organisms, the information obtained through proteomic studies is increasingly complete (Suresh et al., 2005). This new knowledge is related to cell signaling pathways, regulatory protein assemblies, post-translational modifications, as well as other crucial information on the physiological and pathophysiological states of cells and organisms. The term proteome was proposed in 1995 by Wilkins as being the entire content of proteins expressed by a genome. After the euphoria caused by the sequencing of the genomes of several organisms, the scientific community realized that in order to understand the gene function in its fullness, the large-scale study of the expressed proteins was necessary. Among the main techniques used in proteomics are two-dimensional electrophoresis and mass spectrometry. Electrophoresis is a separation technique based on the migration of charged molecules into a solution, depending on the application of an electric field. Protein electrophoresis was first performed in 1937 by Arne Tiselius, who devised a method called free electro- phoresis, which consisted of the decomposition of blood serum into five main protein fractions; this study was important for him to win a Nobel Prize. In the last decades, this technique experienced constant improvements, enabling more precise analyzes, as in the case of denaturing electrophoresis in polyacrylamide gel and two-dimensional electrophoresis.
Polyacrylamide gel electrophoresis in the presence of sodium dodecyl sulfate (SDS) is a widely used method for the analysis of molecular masses of oligomeric proteins. The gel is a matrix consisting of a crosslinked acrylamide polymer of N, N-methyl-bis-acrylamide whose mesh porosity can be chosen. The higher the acrylamide concentration, the smaller the pores of the formed mesh. After the treatment with amphipathic detergent, proteins are applied to the top of a polyacrylamide gel and subjected to an electric current, making them to migrate through the acrylamide mesh towards the direction of the positive pole. Depending on its size, each protein will move differently, with smaller proteins migrating faster, while larger ones will have more difficulty in moving the gel mesh and thus they will move more slowly. When electrophoretic mobility is plotted against the logarithm of known molecular weights of various polypeptide chains (labeling proteins), a straight line is obtained which can be used as the standard for calculating molecular weight of the subunits of the protein of interest.
Mutant study
Genetic variability forms the base in the essence of evolutionary processes and plant breeding; it is essential for the natural and/or artificial selection to be effective (Jennings et al., 1981). In addition to the pre-existing genetic variability in the germplasm, it is possible to add variability through artificial mutations, gene recombination, genetic transformation and soma-clonal mutations. For more than 50 years, different strategies have been used in this regard, and the many rice cultivars have been produced from induced mutations and/or selection of genetic constituents from mutant populations. In many cases, a point mutation can correct or improve some traits, allowing the selection of superior genetic constitutions in the early generations. This strategy was used to change the frequency of some genes that are negligible by the expression of traits in the IAC-24 wheat cultivar, allowing the selection of at least eight lines that maintained the original characteristics and had traits such as the height of plants and tolerance to pathogens (Tulmann-Neto et al., 1995). In triticale, mutation induction was efficient in reducing the height of plants like those produced by artificial crosses (Pandini et al., 1997).
Researchers in Brazil have used 60Co mutation induction to generate genetic variability in the BRS 7 "Taim" rice cultivar, belonging to the modern group (Filipino), characterized by short and erect leaves with average height of about 80 cm, with high tillering capacity and a biological cycle, in the southern part of the state of Rio Grande do Sul; with 125-130 days of emergence, complete maturation and sensitivity to cold, mainly in the reproductive phase. In the M2 generation, 623 mutants were selected for different traits, including the height of plant and cycle (Zimmer et al., 2003). In a study developed by Martins et al. (2005) in which the variability for morphological traits in rice mutants was studied, it was observed that the seed irradiation of the cultivar BRS 7 "Taim" with 60Co was efficient in the generation of mutants for the traits: height of plant, cycle, number of tiller, number of panicles and index of fertile tiller.
Patch-clamp
Some techniques of the study on transporters and other membrane proteins related to the transport of nutrients in plants can be used in parallel with genetic breeding in order to obtain plants with higher nutritional value. The patch-clamp method consists of, a tiny area of the cell membrane (patch) and making recordings of chains that flow through it (Barry and Lynch, 1991). A glass micropipette, filled with a suitable electrolytic solution and with resistance ranging from 4 to 6 MΩ, is pressed against the membrane and melts with it, forming an extremely high resistance and mechanical stability seal. Subsequently, the membrane patch is ruptured by suction with the pipette still sealed to the cell, providing access to the interior of it. A high strength seal is required for two reasons: first, the higher the seal strength, the more complete is the membrane patch insulation with respect to the external solution, and second, a high resistance decreases the chain that can pass between the pipette and the membrane. In this way, all the ions that flow, when the ion channels open, must flow into the pipette. The resulting electric chain, although small, can be measured with an ultra-sensitive amplifier connected to the pipette. Fuchs et al. (2005) confirmed that rice K+ uptake channel OsAKT1 is sensitive to salt stress by the patch-clamp method. They used this technique on rice root protoplasts to identify a K+ inward rectifier with similar channel properties as heterologously expressed OsAKT1. This technique was developed for studies on the behavior of ion channels and presents a great potential to be used in plant breeding research regarding the selection of individuals with a higher number of specific carriers for certain nutrients in root canal tissues. Also, patch-clamp technique can help in the discovery of potential membrane transporters to be inserted in cultivated species, thus improving, the efficiency of the absorption of nutrients even under conditions of less availability in the soil.
Some researchers pointed out the possibility of performing agronomic biofortification, through fertilization with positive effects on the nutritional quality of the food. However, the genetic biofortification together with the agronomic one extends the possibilities of development of new cultivars more efficient in accumulating minerals and vitamins in the edible part. It is necessary to verify the occurrence of variability among the rice genotypes for the Fe, Zn and the B complex vitamins, so that it will be possible to select these materials within breeding programs. The simple selection of these superior genotypes in relation to nutritional quality, even though by means of conventional breeding techniques can have benefits on rice consumption by the human population. With the advent of genetic engineering, following the methods presented so far, it is possible to generate a rice plant that produces high-concentration β-carotene grains. The knowledge of the enzymes involved in the β-carotene biosynthesis was substantial to the success of obtaining this product that can combat problems of nutritional deficiency in low-income populations. As strategies to be adopted in rice breeding, aiming at biofortification of the grain is important in the techniques of genetic engineering and biotechnology. In parallel with the knowledge on genetics, some plant nutrition concepts should be adopted by breeding programs, especially the biomolecular aspects of plant nutrition.
The authors have not declared any conflict of interests.
REFERENCES
|
Andersson MS, Saltzman A, Virk PS, Pfeiffer WH (2017). Progress update: crop development of biofortified staple food crops under HarvestPlus. Afr. J. Food Agric. Nutr. Dev. 17:11905-11935.
Crossref
|
|
|
|
Barry PH, Lynch JW (1991). Liquid junction potentials and small cell effects in patch clamp analysis. J. Membrane Biol. 121:101-117.
Crossref
|
|
|
|
Bekaert S, Storozhenko S, Mehrshahi P, Bennett MJ, Lambert W, Gregory JF 3rd, Schubert K, Hugenholtz J, Van Der Straeten D, Hanson AD (2007). Folate biofortification in food plants. Trends Plant Sci. 13:28-35.
Crossref
|
|
|
|
Beyer P, Al-Babili S, Ye X, Lucca P, Schaub P, Welsch R, Potrykus I (2002). Golden Rice: Introducing the β-carotene biosynthesis pathway into rice endosperm by genetic engineering to defeat vitamin A deficiency. J. Nutr. 132:506-510.
Crossref
|
|
|
|
Bouis HE, Saltzman A (2017). Improving nutrition through biofortification: A review of evidence from HarvestPlus, 2003 through 2016. Glob. Food Secur. 12:49-58.
Crossref
|
|
|
|
Burkhardt PK, Beyer P, Wünn J, Klöti A, Armstrong GA, Schledz M, Lintig J von, Potrykus I (1997). Transgenic rice (Oryza sativa) endosperm expressing daffodil (Narcissus pseudonarcissus) phytoene synthase accumulates phytoene, a key intermediate of provitamin A biosynthesis. Plant J. 11:1071-1078.
Crossref
|
|
|
|
Datta K, Rai M, Parkhi V, Oliva N, Tan J, Datta SK (2006). Improved 'golden' indica rice and transgeneration enhancement of metabolic target products of carotenoids (β-carotene) in transgenic elite (IR64 and BR29) indica 'golden' rice. Curr. Sci. 91:935-939.
|
|
|
|
Datta SK, Datta K, Parkhi V, Rai M, Baisakh N, Sahoo G, Rehana S, Bandyopadhyay A, Alamgir Md, Ali MdS, Abrigo E, Oliva N, Torrizo LC (2007). Golden rice: introgression, breeding, and field evaluation. Euphytica 154:271-278.
Crossref
|
|
|
|
Diaz de la Garza R, Quinilivan EP, Klaus SM, Basset GJ, Gregory JF 3rd, Hanson AD (2004). Folate biofortification in tomatoes by engineering the pteridine branch of folate synthesis. Proc. Natl. Acad. Sci. 101:13720-13725.
Crossref
|
|
|
|
FAO, IFAD, UNICEF, WFP and WHO (2017). The State of Food Security and Nutrition in the World 2017. Building resilience for peace and food security. FAO, Rome, FAO.
View
|
|
|
|
Finley JW (2006). Bioavailability of selenium from foods. Nutr. Rev. 64(3):146-151.
Crossref
|
|
|
|
Forno DA, Yoshida S, Asher CJ (1975). Zinc deficiency in rice. II. Studies on two varieties differing in susceptibility to zinc deficiency. Plant Soil 42:551-563.
Crossref
|
|
|
|
Frossard E, Bucher M, Mächler F, Mozafar A, Hurrell R (2000). Potential for increasing the content and bioavailability of Fe, Zn and Ca in plants for human nutrition. J. Sci. Food Agric. 80:861-879.
Crossref
|
|
|
|
Fuchs I, Stölzle S, Ivashikina N, Hedrich R (2005). Rice K+ uptake channel OsAKT1 is sensitive to salt stress. Planta 221:212-221.
Crossref
|
|
|
|
García MJ, Suárez V, Romera FJ, Alcántara E, Pérez-Vicente RA (2011). New model involving ethylene, nitric oxide and Fe to explain the regulation of Fe-acquisition genes in Strategy I plants. Plant Physiol. Biochem. 49:537-544.
Crossref
|
|
|
|
Godfray HCJ, Beddington JR, Crute IR, Haddad L, Lawrence D, Muir JF, Pretty J, Robinson S, Thomas SM, Toulmin C (2010). Food security: the challenge of feeding 9 billion people. Sci. 327:812-818.
Crossref
|
|
|
|
Goto F, Yoshihara T, Shigemoto N, Toki S, Takaiwa F (1999). Iron fortification of rice seed by the soybean ferritin gene. Nat. Biotechnol. 17:282-291.
Crossref
|
|
|
|
Graham RD, Rosser JM (2000). Carotenoids in staple foods: their potential to improve human nutrition. Food Nutr. Bull. 21(4):404-409.
Crossref
|
|
|
|
Gregorio GB, Htut T, Cabuslay GS (2008). Breeding for micronutrient enriched rice. In. Ba-uelos GS, Lin ZQ (Ed.) Development and use of biofortified agricultural products. CRC Press, Taylor & Francis Group, NY. pp. 171-180.
|
|
|
|
Gregorio G, Senadhira D, Htut H, Graham R (2000). Breeding for trace mineral density in rice. Food Nutr. Bull. 21(4):382-387.
Crossref
|
|
|
|
Impa SM, Gramlich A, Tandy S, Schulin R, Frossard E, Johnson-Beebout S (2013). Internal Zn allocation influences Zn deficiency tolerance and grain Zn loading in rice (Oryza sativa L.). Front Plant Sci. 4:534.
Crossref
|
|
|
|
Jabrin S, Ravanel S, Gambonnet B, Douce R, Rébeillé F (2003). One-carbon metabolism in plants. Regulation of tetrahydrofolate synthesis during germination and seedling development. Plant Physiol. 131:1431-1439.
Crossref
|
|
|
|
Jena KK, Nissila EAJ (2017). Genetic Improvement of Rice (Oryza sativa L.). In: Genetic Improvement of Tropical Crops. Springer, Cham. pp.111-127.
Crossref
|
|
|
|
Jennings PR, Coffman WR, Kauffman HE (1981). Mejoramiento de arroz. Cali: Centro Internacional de Agricultura Tropical. P 237.
|
|
|
|
Khalekuzzaman M, Datta K, Oliva N, Alam MF, Joarder OI, Datta SK (2006). Stable integration, expression and inheritance of the ferritin gene in transgenic elite rice cultivar BR29 with the enhanced iron level in the endosperm. Ind. J. Biotechnol. 5:26-31.
|
|
|
|
Klein TM, Arentzen R, Lewis PA, Fitzpatrick-Mcelligott SP (1992). Transformation of microbes, plants, and animals by particle bombardment. Biotechnology (NY) 10(3):286-291.
Crossref
|
|
|
|
Marschner H (1995). Mineral nutrition of higher plants. (2nd ed.) Academic Press, San Diego. P 889.
|
|
|
|
Martins AF, Zimmer PD, Oliveira AC, Carvalho FIF, Vieira EA, Carvalho MF, Martins L F, Fonseca FS (2005). Variabilidade para caracteres morfológicos em mutantes de arroz. Ciênc. Agrotec. 29(6):1215-1223.
Crossref
|
|
|
|
Mingotte FLC, Gonçalves MHG, Yada MM, Fornasieri Filho D, Lemos LB (2015). Agronomic efficiency and grain quality of upland rice cultivars as a function of nitrogen topdressing. Biosci. J. 31(3):748-758.
Crossref
|
|
|
|
Mingotte FLC, Hanashiro RK, Fornasieri Filho D (2012). Physico-chemical parameters of rice cultivars fertilized with nitrogen. Semina: Ciências Agrárias. 33(1):2605-2618.
|
|
|
|
Mingotte FLC, Hanashiro RK, Fornasieri Filho D (2013). Response of rice cultivars to nitrogen in upland conditions. Rev. Ceres. 60(1):086-095.
|
|
|
|
Murray-Kolb L, Takaiawa F, Goto F, Yoshihara T, Theil E, Beard J (2000). Transgenic rice is a source of iron for iron-depleted rats. J. Nutr. 132(5):957-960.
Crossref
|
|
|
|
Norton GJ, Douglas A, Lahner B, Yakubova E, Guerinot ML, Pinson SRM, Tarpley L, Eizenga GC, McGrath SP, Zhao FJ, Islam MR, Islam S, Duan G, Zhu Y, Salt DE, Meharg AA, Price AH (2014). Genome wide association mapping of grain arsenic, copper, molybdenum and zinc in rice (Oryza sativa L.) grown at four international field sites. PLoS ONE 9:e89685.
Crossref
|
|
|
|
Palmgren MG, Clemens S, Williams LE, Krämer U, Borg S, Schjørring JK, Sanders D (2008). Zinc biofortification of cereals: problems and solutions. Trends Plant Sci. 13:464-473.
Crossref
|
|
|
|
Pandini F, Carvalho FIF, Barbosa Neto JF (1997). Plant height reduction in populations of Triticale (X triticosecale Wittmack) by induced mutations and artificial crosses. Braz. J. Genet. 20(3):483-488.
Crossref
|
|
|
|
Parker RMC, Barnes NM (1999). mRNA: detection by in situ and northern hybridization. Methods Mol. Biol. 106:247-283.
|
|
|
|
Pereira LFP, Vieira LGE (2006). Transformação de plantas. In: Carpentieri-Pipolo V, Garcia JE. Biotecnologia na agricultura: aplicações e biossegurança. Cascavel: COODETEC. P 392.
|
|
|
|
Poletti S, Gruissen W, Sautter C (2004). The nutritional fortification of cereals. Curr. Opin. Biotechnol. 15:162-165.
Crossref
|
|
|
|
Ponnanperuma FN (1972). The Chemical of submerged soils. Adv. Agron. 21:29-96.
Crossref
|
|
|
|
Potrykus I (1991). Gene transfer to plants: assessment of published approaches and results. Annu. Rev. Plant Physiol. Plant Mol. Biol. 42:205-225.
Crossref
|
|
|
|
Sharma P, Aggarwal P, Kaur A (2017). Biofortification: A new approach to eradicate hidden hunger. Food Rev. Int. 33:1-21.
Crossref
|
|
|
|
Southern EM (1975). Detection of specific sequences among DNA fragments separated by gel electrophoresis. J. Mol. Biol. 98:503-517.
Crossref
|
|
|
|
Stangoulis JCR, Huynh BL, Welch RM, Choi EY, Graham RD (2007). Quantitative trait loci for phytate in rice grain and their relationship with grain micronutrient content. Euphytica 154:289-294.
Crossref
|
|
|
|
Storozhenko S, Navarette O, Ravanel S, De Brouwer V, Chaerle P, Zhang GF, Bastien O, Lambert W, Rébeillé F, Der Straeten DV (2007). Cytosolic hydroxymethyldihydropterine pyrophosphokinase/dihydropteroate synthase from Arabidopsis thaliana: a specific role in early development and stress response. J. Biol. Chem. 282: 10749-10761.
Crossref
|
|
|
|
Suresh S, Mohan SS, Mishra G, Hanumanthu GR, Suresh M, Reddy R, Pandey A (2005). Proteomic resources: Integrating biomedical information in humans. Gene 364:13-18.
Crossref
|
|
|
|
Tan J, Baisakh N, Oliva N, Torrizo L, Abrigo E, Datta K, Datta SK (2004). The screening of rice germplasm including those transgenic rice lines which accumulate b-carotene in their polished seeds for their carotenoid profile. Int. J. Food Sci. Technol. 40:563-569.
Crossref
|
|
|
|
Thomas PS (1983). Hybridization of denatured RNA transferred on dotted nitrocellulose paper. Methods Enzymol. 100:255-266.
Crossref
|
|
|
|
Tulmann-Neto A, Camargo CEO, Alves MC, Santos RR, Freitas JG (1995). Indução de mutação visando obtenção de resistência a doenças no cultivar de trigo IAC-24. Pesq. Agropec. Bras. 30(4):497-504.
|
|
|
|
Vasconcelos M, Datta K, Oliva N, Khalekuzzaman M, Torrizo L, Krishnan S, Oliveira M, Goto F, Datta SK (2003). Enhanced iron and zinc accumulation in transgenic rice with the ferritin gene. Plant Sci. 164:371-378.
Crossref
|
|
|
|
Viviani VR (2008). Luciferases de vagalumes: estrutura, função e aplicação em bioanálise e bioimageamento. Biotecnolog. Ciênc. Desenvolv. 37:8-19.
|
|
|
|
Weis JH, Tan SS, Martin BK, Wittwer CT (1992). Detection of rare mRNAs via quantitative RT-PCR. Trends Genet. 8:263-264.
Crossref
|
|
|
|
White PJ, Broadley MR (2005). Biofortifying crops with essential mineral elements. Trends Plant Sci. 10:586-593.
Crossref
|
|
|
|
Whanger PD (2002). Selenocompounds in plants and animals and their biological significance. J. Am. Coll. Nutr. 21:223-232.
Crossref
|
|
|
|
Yang X, Romheld V, Marschner H (1994). Effect of bicarbonate and root growth and accumulation of organic acid in Zn-inefficient and Zn-efficient rice varieties (Oryza sativa L.). Plant Soil 164:1-7.
Crossref
|
|
|
|
Yang YH, Dudoit S, Luu P, Lin DM, Peng V, Ngai J, Speed TP (2002). Normalization for cDNA addressing single and multiple slide systematic variation. Nucleic Acids Res. 30:4-15.
Crossref
|
|
|
|
Zancul MS (2004). Food fortification with iron and vitamin A. Medicina. 37:45-50.
|
|
|
|
Zhang W, Wu R (1988). Efficient regeneration of transgenic plants from rice protoplasts and correctly regulated expression of the foreign gene in the plants. Theor. Appl. Genet. 76:835-840.
Crossref
|
|
|
|
Zimmer PD, Mattos LAT, Oliveira AC, Carvalho FIF, Magalhães Jr A, Kopp MM, Freitas FA (2003). Identification of rice mutants (Oryza sativa L.) for agronomical and root system traits. Rev. Bras. Agrocienc. 9(3):195-199.
|
|
|
|
Zuo Y, Zhang F (2011). Soil and crop management strategies to prevent iron deficiency in crops. Plant Soil. 339:83-95.
Crossref
|