ABSTRACT
Wuchereria bancrofti has been reported to cause 90% of all cases of lymphatic filariasis in Africa. wolbachia endosymbiot infect a wide range of insects and nematodes. This study was conducted in 48 settlements (8 from each division). A total of 2003 mosquitoes was pooled into 120 clusters from the 48 settlements in The Gambia. Polymerase chain reaction (PCR) was used to detect the presence of Wolbachia and filarial nematode and further differentiate Wolbachia into super groups among the positive samples. The level of association between Wolbachia and the genera of anthropophilic mosquitoes in The Gambia was also determined. Microscopic results showed 64.9% Anopheles, 32.0% Culex, 3.0% Aedes. PCR showed that, 34.17% of Wolbachia in the mosquito obtained varied among the three mosquito genera, with highest being among Anopheles. Only Wolbachia super group B was identified in Culex and Aedes. The purpose of this study carried out in The Gambia was to describe the most common mosquito species and to identify, by means of PCR, their association with the Wolbachia bacteria and the presence of nematodes responsible for filiariasis, in order to understand the role that this bacterium plays in the chain of filarial transmission.
Key words: Wuchereria bancrofti, Filariasis, Anopheles gambiae, Culex quinquefasciatus, elephantiasis.
Lymphatic filariasis, typically known as elephantiasis and it is one of the neglected tropical disease caused by a microscopic parasite which can affect man (WHO, 2020). It can disfigure and damage the lymphatic system and can cause abnormal enlargement of body parts, thereby causing grave pain, severe disability and stigma. About 893 million people in 49 countries worldwide are at risk of contracting this disease (WHO, 2020). It is caused by a thread- like filarial worms (microfilariae): Wuchereria bancrofti, Brugia malayi or Brugia timori (Lambert, 2005). The adult worms which could measure up to 4 -12 cm long, house and mature as adults in the body‘s lymph nodes and lymph vessels for a period of 4 to 8 years (Bockarie et al., 2009; Pfarr et al., 2009). Three species that causes lymphatic filariasis, W. bancrofti is most widely spread and is responsible for more than 90% of
infection (Rebollo et al., 2015a). Microfilariae of W. bancrofti exhibit nocturnal periodicity (Nwoke et al., 2010); during the day, they are found in the deep veins and during the night between 10 pm and 4 am, they migrate to the peripheral circulation. This behaviour gives them the chances of being picked up by the night biting vectors during their blood meals (Nwoke et al., 2010). The species of mosquitoes capable of transmitting these filarial nematodes are many but varies between geographical regions. In most urban and semi urban areas, W. bancrofti is vectored mainly by Culex mosquito while in the rural areas, transmission from one person to another is mainly by Anopheles mosquito (Singh et al., 2013). The main vectors in The Gambia are Anopheles gambiae in the rural areas while in the urban areas transmission is by Culex quinquefasciatus. Studies have revealed the presence of an endosymbiont, Wolbachia in arthropods and nematodes (Bouchery et al., 2012). Wolbachia is a Gram-negative intracellular α-proteobacterium belonging to the Order Rickettsiales (Bouchery et al., 2012). The specie Wolbachia pipientis was first identified in Culex mosquito (Hertig and Wolbach, 1924; Porennan et al., 2008). It is classified into sixteen sub groups (A-Q) (Guan et al., 2016). Super group G have been removed because its status is currently not clear (Baldo and Werren, 2007). Of the sixteen sub groups, C and D are commonly found in filarial nematodes (Werren et al., 2008), while A and B are found in mosquitoes (Ravikumar et al., 2011). According to Dyab et al., (2016), Wolbachia are maternally inherited and infect a wide range of insects and nematodes and play an important role in the development and pathogenesis of the filariae Onchocerca volvulus, Brugia malayi and W. bancrofti. Wolbachia as a parasite, relates with its host in a number of ways, some of which are parasitism while others are mutualism. The parasitic relationship includes feminization of genetic males, pathenogenetic induction resulting in the development of unfertilized eggs, the killing of male progeny from infected females and sperm-egg incompatibility (Werren et al., 2008). Wolbachia appear to contribute to the inflammation process which is a major pathological symptom of filarial infections (Dyab et al., 2016). The most common effect of Wolbachia infection in mosquitoes is cytoplasmic incompatibility. Mating of infected male with the uninfected female mosquitoes of the same species, does not result into fertilization (Sinkins, 2004). Wolbachia has the potential to be used as biocontrol in both pest and in biomedical applications (Zabalou et al., 2004; Engelstadter and Hurst, 2009). In 2013, the World Health Organization listed The Gambia as among 73 countries considered endemic for lymphatic filariasis (WHO, 2013) and according to Cano et al. (2014), Global Atlas of Helminths Infection, The Gambia still had a high probability of transmission. Lymphatic filariasis is mainly characterized by the occurrence of inflammatory pathogenesis, and Wolbachia bacterium which is an endosymbiont of filarial nematode appear to contribute to this inflammation (Barton et al., 2010; Bouchery et al., 2013). The endosymbiont Wolbachia could be genetically transformed to modify the disease transmitting abilities of mosquitoes (Yeap et al., 2011; Fraser et al., 2017). It is crucial to know which Wolbachia strains are present in population before releasing engineered infected individuals because pre-existing natural infections can interact with and alter the dynamics of introduced strains (Duron et al., 2010; Atyame et al., 2011). Understanding the role of Wolbachia in filarial transmission can provide a template for breaking the transmission chain of the disease. Effective and efficient surveillance systems need to be put in place for most arthropod-borne diseases across The Gambia (Kargbo and Kuye, 2020). The purpose of this study carried out in The Gambia was to describe the most common mosquito species and to identify, by means of polymerase chain reaction (PCR), their association with the Wolbachia bacteria and the presence of nematodes responsible for filiariasis, in order to understand the role that this bacterium plays in the chain of filarial transmission.
Study area
The Gambia is a West African country that lay between latitude 13° and 14° and longitude 13° and 17°. The country is almost completely surrounded by Senegal except on the western side where it bordered with the Atlantic Ocean. The Gambia is divided into eight Local Government Areas namely: Banjul, Kanifing, Brikama, Mansa Konko, Kerewan, Kuntaur, Janjanbureh and Basse. For the purpose of this research, the former divisions as shown in Figure 1 was used; Banjul, Western, North Bank, Lower River, Central River and Upper River division.
Sample size
The sample size was determined by using the formula
N = Z2 pq/ d2
as described by Thrusfield (2007) based on 95% confidence interval and prevalence of 50%.
Where;
N is the sample size of pools of mosquito (to be calculated)
Z = 1.96 for 95% confidence level
p= 0.5 (Prevalence)
q= Complementary probability (1-0.5)
d= 0.05
N = 1.96×1.96x0.5×0.5/0.05x0.05
N = 0.9604/0.0025
N = 384 pools of mosquitoes
Sampling techniques
Multi stage sampling technique was employed. For each division, 8 settlements were selected through balloting, and for each settlement, 8 compounds were selected through balloting. The compounds selected are the sampling points/pools. This equates to 64 sampling points/pools for each division giving a total of 384 sampling points/pools for the entire country (Table 1).
Collection of mosquito samples
G‘rumba is a name of a container in Jola language made of clay. Jola is a tribe in the Senegambia region and some parts of Guinea Bissau deeply rooted in herbal medicine. The container is mostly used by herbalists in the preparation and storage of concoctions. It has been observed that while in storage, a scourge of mosquitoes seeks shelter and a source of water for drinking in such containers. The container could also be a breeding ground if kept in that position for weeks. It is from this concept; this study formulated a trap for collecting mosquitoes for purpose of this research. The materials used here are buckets and transparent piece of clothes sawed into sacs with a control valve made of rope. In the setup, water is filled into the buckets to one quarter full. Small branches with leaves of shrubs were then put inside to create a bushy environment that can be a temporary home for mosquitoes and other flies within that environment. The harvesting processes involved carefully capping the mouth of the bucket with the mouth of the designed sac such that when the bucket is shaken the mosquitoes will run into the sacs. The sac can then be removed carefully and the valve closes to prevent any trapped mosquito from escaping. The activities of mosquitoes are time bound. Some genera are active during the night while others are active in some parts of the day. At around midday, mosquitoes are generally not active. During such periods, they could be found hanging on walls in dark areas or any other surfaces nearby. In this method of collection, the traps were set to provide such resting platforms for the mosquitoes during such inactive periods of their time. The traps were positioned in baths rooms, verandas, sitting rooms and other immediate environments. The compound owners and the community were sensitized on the whole operation. The selected area for the traps was cleared of any other material that could provide a resting platform for the mosquitoes. The traps were laid from 8 am in the mornings to 2 pm the next day (a period of 30 h). Harvests were made at 2 pm as described above. The live mosquitoes were left in the sacs for 48 h to weaken and then transferred into tubes containing silica in sachets (for preservation) and labelled to indicate division, pool number and date of harvest. Once harvested, the small branches with leaves were discarded and the water poured to avoid the setup being used by mosquitoes for breeding (Sankung et al., unpublished method of trapping mosquitoes). The mosquitoes were then transported to Department of Biochemistry Ahmadu Bello University, Zaria Kaduna State, Nigeria for laboratory analysis (Figure 2).

Morphological identification of the mosquitoes
The mosquitoes were identified per pool into genera level according to Stone et al. (1959) using microscopy at the Department of Veterinary Parasitology and Entomology Laboratory Ahmadu Bello University Zaria. The main taxonomic features used were; body colour, spotted and unspotted wings, length of palps in comparison with their proboscis and presence of black strips on body and legs. The number of each genera of mosquito for each pool were established and placed in tubes and labelled. The sex of the mosquitoes was also determined for every pool based on the presence of bushy feathers around the antenna. For improving the sensitivity and cost effectiveness in the DNA analysis by Polymerase Chain Reaction (PCR), each genera of mosquitoes in the sampling points/pools of each settlement/cluster were pooled together. This translates into 48 clusters of Anopheles genera mosquitoes, 48 clusters of Culex genera mosquitoes and 24 clusters of Aedes genera of mosquitoes. DNA was extracted from 120 clusters of mosquitoes.
Identification of Wolbachia and microfilariae
DNA extraction
Dry mosquito samples by cluster were homogenized by grinding into powder in eppendorf tubes (one for each cluster) using fresh micro pestle for each cluster. 100 µl of distilled water was added to each of the samples with less than 5 mosquitoes while 200 µl of distilled water was added to samples with more than 5 mosquitoes. The samples were briefly vortexed to distribute the particles evenly. Quick-DNATM miniprep plus kit (Zymo) was used with strict adherence to manufacturer instructions (Vanek et al., 2011).
DNA quantification
The DNA samples were removed from the freezer, thawed and briefly vortexed. The lower and the upper pedestals of the Nano drop Spectrophotometer (Denovix DS- 11+) were cleaned and blanked with 2 µl of deionized water. The samples were then run one after the other with intermittent cleaning of the lower and the upper pedestals with tissue paper after every sample run. The concentration of the DNA and the absorbance of 260/280 ratio (for purity) were recorded.
Identification of Wolbachia DNA using 16SrDNA and wsp gene
Detection using 16S rDNA
To determine the presence of Wolbachia DNA, PCR technique was employed using 16S Wolb F and 16S Wolb R (forward and reverse primers) specific primers: 16S rDNA (W-Specf 5‘-CATACCTATTCGAAGGGATAG-3‘, W-Specr 5‘-AGCTTCGAGTGAAACCAATTC-3‘) (Baldini et al., 2014). The amplification were carried out with a Gradient Thermo Cycler and the reaction mixture for each of the 120 cluster sample consisted of 4 µl of DNA template, 2.5 µl of 10X buffer, 2.5 µl of 2.5 mM (each) dNTPs, 0.35 µl of 20 µM each of forward and reverse primers (16SWolbF/16SWolbR), 0.25 µl of 5U/µl Taq DNA polymerase and a volume of 15.05 µl DNAse free water to make a final reaction volume of 25 µl. The amplification reaction protocol used was as follows initially of 2 min at 95°C then 40 cycles of 30 s at 95°C, annealing at 55°C for 30 s and extension at 72°C for 30 s and a final extension of 3 min at 72°C with an expected ban size of 438 bp (Baldini et al., 2014; Dyab et al., 2016).
Detection of Wolbachia super group a using wsp136F and wsp691R
All positive samples from 16SrDNA screening were subjected to wsp136F/wsp691R screening. The primer sequence are wsp136F 5‘TGAAATTTTACCTCTTTTC 3 and wsp691R 5‘AAAAATTAAACGCTACTCCA 3‘. These primers are specific to Wolbachia super group A. PCR amplification were carried out for all the positive samples for 16SrDNA. A total reaction volume for a single reaction used was 25 µl. The mix contained 4 µl of DNA template, 2.5 µl of 10x reaction buffer, 2.5 µl of dNTPs mix, 0.35 µl each of forward (wsp136f) and reverse (wsp691r) primer, 0.127µl of 5U/µl Taq polymerase and 15.175 µl of DNAse free water. The PCR reaction conditions were as follows; 1 cycle of initial denaturing for 1 min at 94°C followed by 35 cycles of 15 s at 94°C of denaturing, 30 s at 55°C of annealing, extension at 72°C for 1 min and a final extension for 7 min at 72°C with an expected amplicon size of 556 bp (Nugapola et al., 2017).
Detection of Wolbachia super group B using wsp81F and wsp522R
PCR amplification was carried out for all the positive samples for 16srDNA using Forward primer wsp81F 5‘TGGTCCAATAAGTGATGAAGAAC 3‘and a reverse primer wsp522R 5‘ ACCAGCTTTTGCTTGATA 3‘. These primers are specific to Wolbachia super group B. A total reaction volume for a single reaction used was 25µl. The mixture contained 4µl of DNA template, 2.5 µl of 10x reaction buffer, 2.5 µl of dNTPs mix, 0.35 µl each of forward (wsp136f) and reverse (wsp691r) primer, 0.127 µl of 5 U/µl Taq polymerase and 15.175 µl of DNAse free water. The PCR reaction conditions were as follows; initial denaturing of 1 cycle of 1 min at 94°C followed by 35 cycles of denaturing at 94°C for 15 s, 30 s at 55°C of annealing, extension at 72°C for 1 min and a final extension for 7 min at 72°C with an expected amplicon size of 442 bp (Nugapola et al., 2017).
Identification of microfilariae specie
Wuchereria bancrofti
The sspI gene is a repeat sequence which is a signature for W. bancrofti. PCR with specific primers for the SspI gene were used to detect the presence of W. bancrofti in the sample. The forward primer for the SspI gene of W. bancrofti was sspI F 5‘-CGT GAT GGC ATC AAAGTA GGG-3‘, and the reverse primer for the SspI gene was SspIR 5‘-CCC TCA CTT ACC ATA AGA CAAC-3‘. The PCR amplification for 48 cluster samples were carried out in a gradient thermal cycler. A total reaction volume of a single reaction of 25 µl, contained 4 µl of DNA template, 2.5 µl of 10x reaction buffer, 2.5µl of dNTPs mix, 0.35 µl each of forward and reverse primer, 0.127 µl of 5 U/µl Taq polymerase and 15.175 µl of DNAse free water. The PCR reaction conditions were as follows: initial denaturing at 95°C for 5 min followed by 38 cycles of 1min at 94°C denaturing, 1 min at 56°C of annealing, 1 min at 72°C of extension and a final extension at 72°C for 10 min (Nugapola et al., 2017).
Brugia malayi
The HhaI gene in Brugia malayi is a tandem repeat sequence of about 320 bp. The HhaI gene is the target in the detection of Brugia malayi. The forward primer used was HhaI F 5‘-GCG CAT AAA TTT ATC AGC-3‘and the reverse primer was HhaI R 5‘- GCG CAA AAC TTA ATTACA AAA GC-3‘. The amplification of the gene was carried out using a Gradient Thermal cycler (Rebollo et al., 2015b). A total reaction volume of each of the 73 cluster samples used was 25 µl. This contained 2 µl of DNA sample, 2.5 µl of 10 x reaction buffers, 2.5 µl of dNTPs mix, 0.35 µl of each of forward and reverse primers, 0.125 µl of 5 U/µl of Taq polymerase and 15.175 µl of DNase free water. The reaction protocol was as follows; 95°C for 5 min, 38 cycles of 30 s at 95°C, 30 s at 56°C 30 s at 72°C, 5 min at 72°C, with an expected product size of 320 bp. The products from the nested PCR were subjected to gel electrophoresis (35 min at 100V) in 1% agarose gel stained with Ethidium and visualized using Chem-Doc Imaging System (BIO-RAD).
Statistical analysis
The association between Wolbachia and different genera of mosquitoes were analysed using bivariate fit of quadratic regression of Statistical Analysis System (SAS 9.4). Relationship between variables was determined using correlation analysis. Charts were fitted in Microsoft Excel. Significant association was set at p < 0.05.
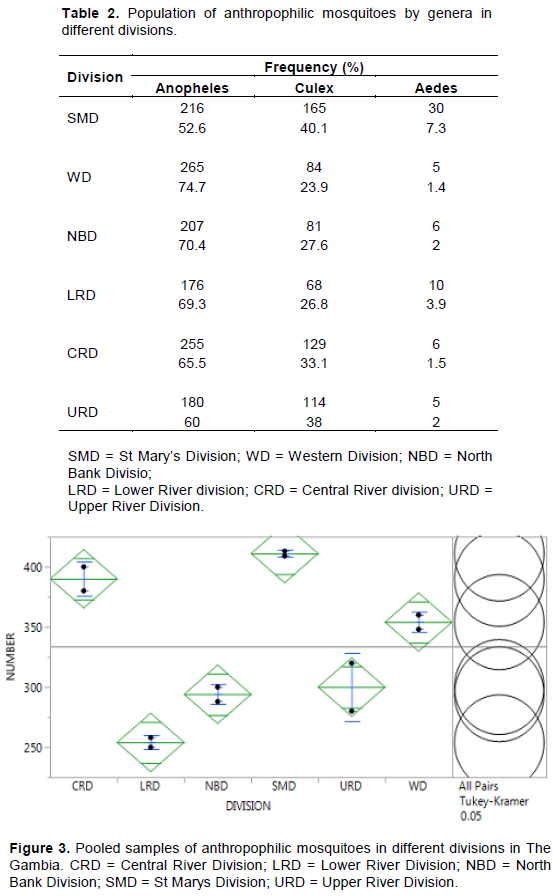
Distribution of Anthropophilic Mosquito Genera in The Gambia. A total of 2003 anthropophilic mosquitoes from six divisions in The Gambia were classified into genera by microscopy method (magnification X10). Three genera of anthropophilic mosquitoes were identified as Anopheles, Culex and Aedes. Mansonia genera of mosquito was not detected in the total anthropophilic mosquitoes collected (Table 2). A total of 65.0% of the anthropophilic mosquitoes in The Gambia are Anopheles genera, 31.9% are Culex genera, while 3.1% belong to Aedes genera. None of the anthropophilic mosquitoes belongs to the Mansonia mosquito genera. The distribution pattern of the genera was similar across the six divisions. Figure 3 shows the pooled samples of anthropophilic mosquitoes in different divisions in The Gambia. There was significant differences (p<0.05) among the mosquito genera across the division. SMD and CRD had the highest population of mosquitoes which was highly significant (p<0.05) different from those caught in the other regions.
Sexual dimorphism of anthropophilic mosquito genera
Sexual dimorphism of 2003 anthropophilic mosquito from the six divisions in The Gambia were determined by microscopic technique, of which 82.1% were female while 13.0% were males. (Table 3) There is a significant different (p<0.05) in sexual dimorphism among anthropophilic mosquitoes in The Gambia.
Detection of Wolbachia and classification into super group
Results of the DNA analysis from 120 clusters of mosquitoes for presence of Wolbachia and super group using 16SrDNA and wsp primers are presented in Figure 4 and 5. The results indicated a 34.17% incidence (Table 4) of Wolbachia in anthropophilic mosquitoes in The Gambia. All the three genera of mosquitoes were infected with Wolbachia (Figure 4). A higher proportion of the Anopheles harboured Wolbachia (50%), while Aedes had a significantly lower rate. The Wolbachia detected in Culex and Aedes belong to Super group B (Figure 5). Though Wolbachia was detected in Anopheles genera using 16SrDNA, the super group could not be determined using wsp primers. This was due to the insensitivity of wsp primers in detecting Wolbachia in Anopheles. However, no filiarial nematode was detected in mosquitoes as seen Figure 6.
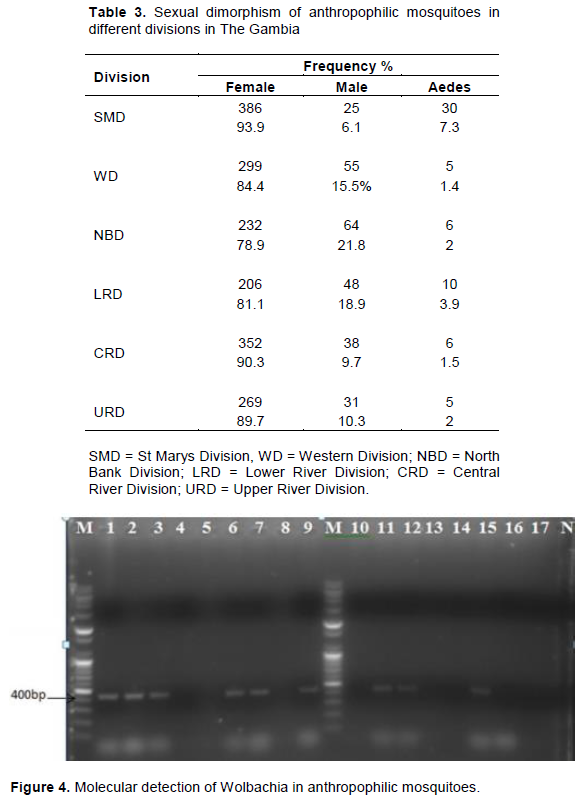
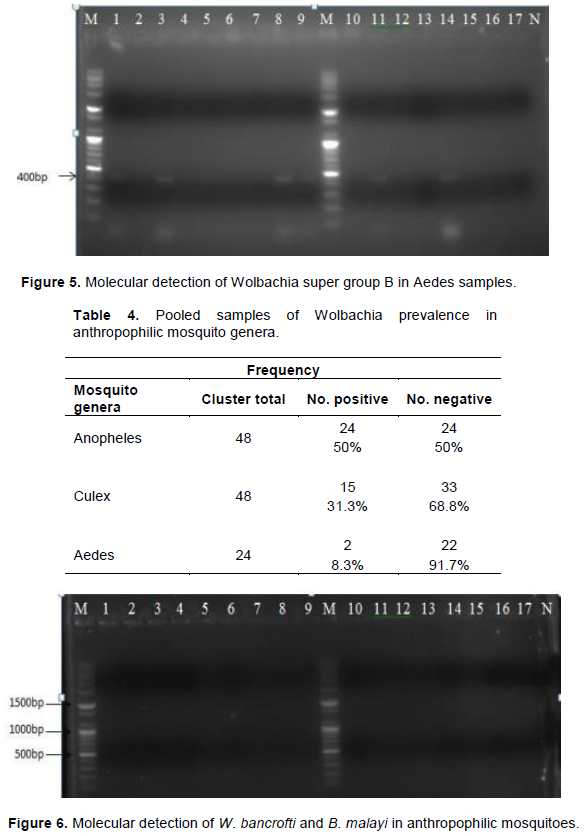
Wolbachia in anthropophilic mosquitoes in The Gambia. Lane M is the DNA marker. Lane N was the negative controls, that is: PCR reaction without gDNA. Samples from mosquitoes are represented in the following lane. PCR amplification of the 16SrDNA (438 bp) of Wolbachia in anthropophilic mosquitoes Lanes 1, 2, 3, 6 and 7 were positive for Anopheles samples. Lanes 9, 11, 12 and 15 was positive for Culex samples (Baldini et al., 2014; Dyab et al., 2016) (Figure 5). Wolbachia in anthropophilic mosquitoes in The Gambia. Lane M is the DNA marker. Lane N was the negative controls, that is: PCR reaction without gDNA. Samples from mosquitoes are represented in the following lane. Lane M is the DNA marker. Lane N was the negative controls, that is: PCR reaction without gDNA. Samples from mosquitoes are represented in the following lane. Lanes 1, 3, 6, 7, 8, 9, 11, 12 was positive for Wolbachia super group B in Culex samples Lane 14 was positive for Wolbachia super group B in Aedes samples. PCR amplification of the wsp (442 bp) for Wolbachia Super group B in anthropophilic Mosquitoes. This size of the signal were calculated by comparing its mobility to that of the standards’ bands in the marker lane as demonstrated by (Nugapola et al., 2017) (Table 5).
Wolbachia – mosquito association
The association of Wolbachia and anthropophilic mosquito’s genera in The Gambia was analysed using bivariate fit of quadratic regression of statistical analysis system (SAS 9.4). The analysis in Table 6 shows that Wolbachia has high and significant association (p<0.05) with Culex. The association between Wolbachia and Culex was strong and positive (r=0.79) with coefficient of determination of 63%. Association of both Anopheles and Aedes with Wolbachia is not significant (p>0.05), though the correlation was high between Wolbachia and Anopheles (r=0.6), the correlation between Wolbachia and Aedes is low (0.61).
Screening for the presence of W. Bancrofti and B. Malayi DNA in anthropophilic mosquitoes.
Results of the screening of the DNA of the mosquitoes for filarial nematode using ssp1 and HhaI gene primers (Figure 6) indicated that there were no nematode in the sampled mosquitoes Both W. bancrofti and B. malayi were absent in the mosquitoes in the six divisions of The Gambia.
Wolbachia in anthropophilic mosquitoes in The Gambia. Lane M is the DNA marker. Lane N was the negative controls, that is: PCR reaction without gDNA. Samples from mosquitoes are represented in the following lane. Lanes 1, 2, 3,4,5,6,7,8,9 were all negative for Anopheles samples, Lanes 10,11,12,13 and 14 were negative for Culex samples Lanes 15, 16 and 17 were all negative for Aedes samples. PCR amplification of the sspI (188 bp) and hhaI (320 bp) of W. bancrofti and B. malayi in anthropophilic mosquitoes Nugapola et al., (2017).
Various genera of mosquitoes in The Gambia are distributed across all the divisions of the country, with the exception of Mansonia genera which was not detected among the anthropophilic family of mosquitoes sampled. Availability of fresh, clean or turbid water plays an important role in the distribution of mosquitoes. Fresh water body from rivers or ponds constitutes the highest environments for mosquito breeding sites. Although SMD and WD lacked with fresh river waters, urbanization has played a major role in providing mosquito breeding grounds through numerous sewages and waste water bodies produced daily by human activities (M’koumfida et al., 2018). In CRD and URD, the high population could be attributed to the existence of fresh water bodies from the river which could provide a good breeding grounds for the mosquitoes. NBD and LRD registered the lowest in anthropophilic mosquito population compare to the other divisions. These two divisions characterized by sparsely distributed populations produces less sewages and waste water bodies couple with salty river waters reduces the chances of mosquito breeding compared to other divisions. The mosquito population in these two divisions are thus hampered by lack of adequate fresh water bodies all year round. The high proportion of Anopheles genera among mosquitoes in The Gambia is not unexpected. Anopheles is not restricted to clearly defined habitats for breeding. The genera have the potentials to breed in all open water bodies (Laporta et al., 2011). The lower proportion of Anopheles in SM when compared to other divisions, could be attributed to the fact that SMD is the most highly dense in human population with the highest industrial activities. SMD has also been designated as the most polluted divisions. Anopheles mosquitoes generally prefer unpolluted water bodies for breeding (Geissbühler et al., 2007; Laporta et al., 2011). It has been reported that pollution as a result of urbanization has eliminated certain species of Anopheles from urban centres (Geissbühler et al., 2007; Laporta et al., 2011). The least populated among the three existing genera in anthropophilic mosquitoes is the Aedes genera. Aedes genera generally breeds in artificial containers such as pots, tyres, open barrels containing clean water mainly of rain or natural containers of rain water such as holes in trees (Paupy et al., 2009; Nazri et al.,2013). The increased awareness in the fight against mosquito borne diseases which have resulted in the cleaning of the environment, by collecting discarded pots, tyres or any container which could hold water for mosquito breeding and the decline in amount of rain water received in recent times might have contributed to the low occurrence of Aedes mosquito genera in The Gambia. Difference in sexual dimorphism among anthropophilic mosquitoes in The Gambia is significantly higher in SMD, WD, CRD and URD where Wolbachia infection was evident in this study. The relationship between these two factors (mosquito sex and Wolbachia) has been explained by several researchers. Wolbachia induces cytoplasmic incompatibility in arthropods and distort the sex ratio (Bordenstein et al., 2001; Charlat et al., 2006; Telschow et al., 2007). Through cytoplasmic incompatibility Wolbachia modifies the spermatozoa such that male mosquitoes dies early in embryogenesis and also Wolbachia was also reported to have the potentials to feminize male mosquitoes during early embryogenesis. The significant differences in sexual dimorphism observed in this study may be attributed to the presence of Wolbachia in anthropophilic mosquito populations. The three genera of mosquitoes; Anopheles, Culex and Aedes among anthropophilic mosquitoes in The Gambia are infected with Wolbachia. The incidence of infection is greater in Anopheles than Culex and Aedes. Although the mosquito-Wolbachia association is significant only in Culex, only the super group B of Wolbachia was identified in Culex and Aedes anthropophilic mosquitoes in The Gambia. The super group in Anopheles could not be detected by wsp primers in this study as also reported by (Kittayapong et al., 2000; Wiwatanaratanaburt, 2013; Nugapola-nalaka et al., 2017). The insensitivity of wsp primers in detecting Wolbachia in Anopheles mosquitoes could be attributed to some form of mutation in the surface protein gene in Wolbachia. The inability of wsp primers in detecting Wolbachia has previously led to the conclusion that Anopheles genera are not naturally infected with Wolbachia (Nugapola et al., 2017). This conclusion spurred scientists in the World to attempt introducing Wolbachia artificially into natural population of Anopheles. However, screening with 16srDNA has now revealed that Anopheles mosquito can also harbour Wolbachia. The super group of the Wolbachia in Anopheles will be better determined by sequencing. None of the 120 clusters of anthropophilic mosquitoes (comprising 2003 mosquitoes) was positive for W. bancrofti and B. malayi, suggesting absence transmission of lymphatic filariasis in The Gambia at the sampling time. This finding is in agreement with the conclusion of Rebollo et al. (2015a) who stated that there is no longer active filarial transmission in the country as at the time of their study. It is probable that the high incidence of Wolbachia infection in anthropophilic mosquitoes could have contributed to this. This assumption is further strengthened by the very high dimorphic difference among the mosquito sexes.
Anopheles mosquitoes constitute the predominant anthropophilic mosquito genera in The Gambia. A high proportion of anthropophilic mosquitoes in the country are infected with Wolbachia, super group B, particularly in Culex and Aedes. W. bancrofti and B. malayi DNA were not detected in anthropophilic mosquitoes in The Gambia and hence no active transmission of lymphatic filariasis in the Gambia exist at the time of this study in The Gambia.
The authors have not declared any conflict of interests.
This work was funded by the Africa Centre of Excellence for Neglected Tropical disease and forensic Biotechnology, Ahmadu Bello University, Zaria, Nigeria and The Africa Centre of Excellence Project Team, The Gambia. Finally, we thank Dr. Idowu Aimola and the Late Dr. T.T Gbem for their helpful comments and assistance in the course of this work.
REFERENCES
|
Atyame CM, Duron O, Tortosa P, Pasteur N, Fort P, Well M (2011). Multiple Wolbachia determinant control the evolution of cytoplasmic incompatibilities in Culex pipiens mosquito populations. Molecular Ecology 20(2):286-298.
Crossref
|
|
|
|
Baldini FN, Jullen P, Perrine MW, Robert S, Roch KD, Abdoulaye D, Elena A, Levashina A, Flaminia C (2014). Evidence of natural Wolbachia infections in field populations of Anopheles gambiae. Nature Communication 6(5):3985.
Crossref
|
|
|
|
|
Baldo L, Werren H (2007). Revising Wolbachia super group typing based on wsp: Spurious Lineages and discordance with MLST. Current Microbiology 55:367-373.
Crossref
|
|
|
|
|
Barton ES, Mark JT, Jeremy MF (2010). The Wolbachia endosymbiont as anti-filarial nematode target. Journal of Symbiosis 51:55-65.
Crossref
|
|
|
|
|
Bockarie MJ, Taylor MJ, Gyapong JO (2009). Current practices in the management of lymphatic filariasis. Expert Review Anti-infection 7:595-605
Crossref
|
|
|
|
|
Bordenstein SR, Patrick O, Werren JH (2001). Wolbachia-induced incompatibility precedes other hybrid incompatibilities in Masonia. Nature 409(6821):675-677.
Crossref
|
|
|
|
|
Bouchery T, Lefoulon E, Karadjian G, Nieguitsila A, Martin C (2013). The symbiotic role of Wolbachia in Onchocercidae and its impact on filariasis. Elsevier 9(2):131-140.
Crossref
|
|
|
|
|
Bouchery T, Lefoulo E, Karadjia G, Nieguitsila A, Martin C (2012). The symbiotic role of Wolbachiain Onchocercidae and its impact on filariasis. Clinical Microbiology and Infection 19(2):131-40.
Crossref
|
|
|
|
|
Cano J, Maria P, Golding N, Pullan RL, Crellen T, Soler A (2014). The global distribution and transmission limits of lymphatic filariasis: Past and present. Parasites and Vectors 7:466.
Crossref
|
|
|
|
|
Charlat S, Reuter M, Dyson EA, Hornett EA, Duplouy AMR, Davies N, Roderick G, Wedell N, Hurst GDD (2006). Male-killing bacteria trigger a cycle of increasing male fatigue and female promiscuity. Journal of Current Biology 17(3):273-277.
Crossref
|
|
|
|
|
Duron O, Raymond M, Weill M (2010). Many compatible Wolbachia strains coexist within natural populations of Culex pipiens mosquito. Heredity 106(6):986-993.
Crossref
|
|
|
|
|
Dyab AK, Lamia AG, Abeer EM, Yasser M (2016). Finding Wolbachiain Filarial larvae and Culicidae Mosquitoes in Upper Egypt Governorate. Korean Journal of Parasitology 54(3):265-272.
Crossref
|
|
|
|
|
Engelstadter J, Hurst GDD (2009). The ecology and evolution of microbes that manipulates host reproduction. Annual. Review of Ecology. Evolution and Systematic 40:127-149.
Crossref
|
|
|
|
|
Fraser E, Bruyne T, Omaetxe I, Stepnell J, Burns L, Flores A, O'Neill L (2017). Novel Wolbachia Trans-infected Aedes aegypti mosquitoes possess diverse fitness and vector competence phenotype. Journal of Arthropods 5(1):211-216.
|
|
|
|
|
Geissbühler Y, Chaki P, Emidi B, Govella NJ, Shirima R, Mayagaya V, Mtasiwa D, Mshinda H, Fillinger U, Lindsay SW, Kannady K, de Castro MC, Tanner M, Killeen GF (2007). Interdependence of domestic malaria prevention measures and mosquito-human interactions in urban Dar es Salaam, Tanzania. Malaria Journal 6:126.
Crossref
|
|
|
|
|
Guan HW, Ling-Yi J, Jin H X, Da-Wei H (2016). Discovery of a new Wolbachia super group in cave spider species and the lateral transfer of phage WO among distant hosts. Journal of Infection, Genetics and Evolution 41:1-7.
Crossref
|
|
|
|
|
Hertig M, Wolbach SB (1924). Studies on Rickettsia-like microorganisms in insects. Journal of Medical Research 44:329-374.
|
|
|
|
|
Kargbo A, Kuye RA (2020). Epidemiology of tsetse flies in the transmission of trypanosomiasis: Technical review of The Gambia experience. International Journal of Biological Science and Chemical Science 14(3):1093-1102.
Crossref
|
|
|
|
|
Kittayapong P, Baisley KJ, Baimai V, O'Neill SL (2000). Distribution and diversity of Wolbachia infection in southeast Asian mosquitoes (Diptera: Culicidae). Journal of Medical Entomology 37(3):340-345.
Crossref
|
|
|
|
|
Lambert SWJ (2005). Lymphatic filariasis: Transmission, Treatment and Elimination. ISBN 10: 90-9019949-7.
|
|
|
|
|
Laporta Z, Ramuo G, Riburo C, Sallum AM (2011). Habitat suitability of Anopheles vector species and association with human malaria in the Atlantic forest in South-eastern Brazil. Memorias do Instituto Oswaldo Cruz 106(1):111-119.
Crossref
|
|
|
|
|
M'koumfida B, Yaffa S, Bah A (2018). The impact of saline-water intrusion on the livelihoods of Gambian Rice Growing Farmers. Research and Review: Journal of Ecology and Environmental Science 6(1):January- March 2018.
|
|
|
|
|
Nazri CD, Abu-Hassan A, Rodziah I (2013). Habitat characterization of Aedes specie breeding in Urban Hotspot Area. Elsevier 85:100-109.
Crossref
|
|
|
|
|
Nugapola-Nalaka NWP, Priyanka WA, De-Silva P, Parakrama-Karunaratne SHP (2017), Distribution and phylogeny of Wolbachia strains in wild mosquito population in Sri Lanka. Parasites and Vectors 10:230.
Crossref
|
|
|
|
|
Nwoke BEB, Nwoke EA, Nkaga CN, Nwachukwu MI (2010). Epidemiological characteristics of Bancroftian filariasis and the Nigerian environment. Journal of Public Health and Epidemiology 2(6):113-117.
|
|
|
|
|
Paupy C, Delatte H, Bagny L, Corbel V, Frontenille D (2009). Aedes albopictus, an arbovirus vector from the darkness to light. Elsevier 11:1177-1185.
Crossref
|
|
|
|
|
Pfarr KM, Debrah AY, Specht S, Hoerauf A (2009). Filariasis and lyphoedema: Parasite Immunology 31:664-672.
Crossref
|
|
|
|
|
Porennan LT, Keddie BA, Braig HR, Harris HL (2008). The endosymbiont Wolbachia pipiens induces the expression of host antioxidant proteins in an Aedes albopictus cell line. PLoS ONE 3(5):e2083.
Crossref
|
|
|
|
|
Ravikumar H, Prakash BM, Sampathicumar S, Puttaraju HP (2011). Molecular subgrouping of Wolbachia and bacteriophage WO infection among some Indian Drosophila specie. Journal of Genetic 90:507-510.
Crossref
|
|
|
|
|
Rebollo MP, Sambou SM, Thomas B, Biritwum N-K, Jaye MC, Kelly-Hope L, Escalada AG, Molyneux DH, Bockarie MJ (2015a). Elimination of Lymphatic Filariasis in The Gambia. PLOS Neglected Tropical Diseases 9(3):e0003642.
Crossref
|
|
|
|
|
Rebollo MP, Sime H, Assefa A, Cano J, Deribe K, Gonzalez-Escalada A, Shafi O, Davey G, Brooker SJ, Kebede A, Bockarie MJ (2015b). Shrinking the lymphatic filariasis map of Ethiopia: reassessing the population at risk through nationwide mapping. PLoS Neglected Tropical Disease 9(11):e0004172.
Crossref
|
|
|
|
|
Singh G, Raksha A, Urhekar D (2013). Advanced Techniques for Detection of Filariasis: A Review. Journal of Clinical and Diagnostic Research 8(12):FC05-FC08.
|
|
|
|
|
Sinkins SP (2004). Wolbachia and cytoplasmic incompatibility in mosquitoes. Insect Biochemistry and Molecular Biology 34(7):723-729.
Crossref
|
|
|
|
|
Stone A, Knight KL, Starcke H (1959). A synoptic catalog of the mosquitoes of the World (Diptera, Culicidae, Culicini). Thomas Say Foundation 6:1-358.
|
|
|
|
|
Telschow A, Flor M, Kobayashi Y, Hammerstein P, Werren JH (2007). Rees, Mark, ed. "Wolbachia-Induced Unidirectional Cytoplasmic Incompatibility and Speciation. PLoS ONE 2(8):e701.
Crossref
|
|
|
|
|
Thrusfield MV (2007). Veterinary epidemiology. 3rd edition. Oxford Blackwell 610p.
|
|
|
|
|
Vanek D, Silerova M, Urbanova V, Saskova L, Dusbska J, Beran M (2011). Genomic DNA extraction protocols for bone samples: The comparison of Qiagen and Zymo Research spin columns, Forensic Science International: Genetics Supplement 3(1):e397-e398.
Crossref
|
|
|
|
|
Werren JH, Baldo L, Clack M (2008). Wolbachia: Master manipulators of invertebrate biology. Nature Review Microbiology 6(10):51741-51744
Crossref
|
|
|
|
|
WHO (2013). Lymphatic filariasis: Managing morbidity and preventing disability. World Health Organization Global Programme to Eliminate Lymphatic Filariasis. SSBN: 978 92 4 150529 1. 1-20.
|
|
|
|
|
WHO (2020). Lymphatic filariasis, Key facts, 2nd March, 2020.
|
|
|
|
|
Wiwatanaratanaburt I (2013). Geographic distribution of Wolbachial infections in mosquitoes from Thailand. Journal of Invertebrate Pathology 114:337-340.
Crossref
|
|
|
|
|
Yeap HL, Mee P, Walker T, Weeks AR, Neill SL, Johnson P, Ritchie SA, Richardson KM, Doig C, Endersby NM, Hoffmann AA (2011). Dynamics of the popcorn 'Wolbachia infection in outbred Aedes aegypti informs prospects for mosquito vector control. Journal of Genetics 187:583-595.
Crossref
|
|
|
|
|
Zabalou S, Riegler M, Theodorakopoulou M, Stauffer C, Savakis C, Bourtzis K (2004). Wolbachia induced cytoplasmic incompatibility as a means of insect pest population control. Proceedings of National Academy of Science United States of America 19:101(42):15042-15045.
Crossref
|
|