ABSTRACT
Previous proteomic studies have shown maize cob tissue to have an essential role in pathogen defense. Currently, there are no studies published regarding neither maize cob metabolomic profiles nor an accepted method of metabolite extraction from cob. This study assesses the reproducibility of metabolite extraction from the cobs of fungal pathogen -resistant and -susceptible cultivars. Hand grinding, mechanical ball-milling and adapted focused acoustics methods of tissue homogenization were preformed and examined via Liquid Chromatography-Mass Spectrometry (LC-MS). Among tested methods, the manual grinding with a mortar and pestle was found to be the most reproducible and efficient. All methods showed good reproducibility within but provided statistically different sets of metabolite features when compared across methods. A suitable extraction method for future cob metabolomics experiments was ascertained, however careful results validation will be needed to distinguish between true biological phenomenon and artifacts rising from a methodology bias. For the first time, maize cob metabolites have been successfully extracted and detected, establishing a reproducible method for further metabolomic profiling of cob defense metabolites.
Key words: Liquid Chromatography-Mass Spectrometry (LC-MS), metabolomics, Aspergillus flavus, homogenization, extraction, grinding.
In maize (Zea mays L.), the cob provides mechanical support to kernels in addition to transporting essential nutrients and water. Previous studies have shown that cob tissue plays an essential role in both facilitating and limiting the spread of the fungal pathogen Aspergillus flavus which causes ear rot and produces the carcinogenetic metabolite aflatoxin B1 (Alfaro, 2000; Magbanua et al., 2013). Previous proteomic analysis of cob tissue revealed the presence of constitutive and fungal-induced defense proteins (Pechanova and Pechan, 2015; Pechanova et al., 2011), suggesting defense metabolites in the parent cob tissue are linked to the defense response in the developing kernel. To date, no metabolomic profiling studies for cob have been published in the literature. This study provides the initial step towards a maize cob global metabolomic profiling study by establishing the impacts of different types of tissue disruption and homogenization on the reproducibility of features detected in metabolomics analysis via Liquid Chromatography-Mass Spectrometry (LC-MS).
Unlike soft tissues of maize, cob is very rigid and presents a challenge as it ages and becomes hardened. Compared to kernel or leaf tissues, cob requires an extensive force to be ground into the fine powder needed for metabolite extraction. No protocol for extraction of metabolites from maize cob tissue has been established in the literature. It has been previously shown that sample preparation and extraction of plant tissues impact the reproducibility of metabolites detected via LC-MS (de Souza et al., 2019; Kim and Verpoorte, 2010; Tugizimana et al., 2018). Thus, this study investigates the reproducibility and efficiency of metabolite extractions from maize cob using three different grinding techniques as an initial step. Methods differ by modes of tissue homogenization, manual grinding, mechanical cryo-grinding, and tissue disruption via Adaptive Focused Acoustic (AFA) technology; but are identical during metabolite elution phase. Mortar and pestle are commonly used for grinding soft tissues into a powder but is labor-intensive and not well suited for callous tissue. The ball mill grinder offers a fast, automated approach to homogenization. The Covaris S220 acoustic focused-ultrasonicator (AFA) was chosen because of its effective cellular disruption previously observed in proteomics.
The A. flavus resistant Mp313E (Scott and Zummo, 1990)and susceptible B73 (Russell, 1972)maize cultivars were chosen as study subjects, due to their availability and connection to previous proteomic studies (Pechanova et al., 2011). Metabolites from three technical replicates for both cultivars were extracted using three different grinding methods and subjected to LC-MS analysis. Raw data files were processed by the Sieve software (ThermoFisher Scientific, Waltham, MA). The Principal Component Analysis (PCA) showed the similarities and differences between genotypes, replicates, and extraction methods. Venn diagrams were created to evaluate method efficiency via detection of unique and overlapping features produced by the three grinding methods for both genotypes.
Metabolite extraction methods
Since metabolites are in flux within a plant system, extreme care was taken to minimize perturbation and/or degradation. Maize plants were field grown in a randomized complete block design with 20 plants per row at Mississippi State University R. R. Foil Plant Science Research Center. Inbreeds were matched as per maturity for each genotype by harvesting ears at 33 days after mid-silk (at least 50 % of the plants in the block had silk showing in the primary ear).
The healthy primary maize ears were harvested from the stalk and were immediately placed on ice during transportation from the field to the laboratory. All kernels, silks, and piths were removed from the cob and a 3.5 cm section of cob was harvested above and below the centermost point. The cob section was then sliced into 1 mm thick disks before being diced. The chopped cob tissue was then flash-frozen in liquid nitrogen, to minimize metabolites degradation (Jones and Kinghorn, 2006), and stored at -80°C until extraction. Six biological replicate tissue samples from each genotype were pooled and split into three technical replicates. When weighing samples, the tubes of stock tissue were removed from cold storage and placed into a liquid nitrogen bath before and after aliquots were measured. Metabolites were extracted by the previously published protocol (Kim and Verpoorte, 2010)but diverse in modes of tissue homogenization. Cob tissue was disrupted to release metabolites by: 1. manual grinding with a mortar and pestle; 2. mechanical grinding; and 3. Adapted Focused Acoustics technology.
Manual grinding (MG)
Diced cob sample (1.5 g) was placed in a nitrogen chilled mortar. The rigid tissue was vigorously ground by a nitrogen chilled pestle for 8-10 min until only a fine powder was obtained. A paper towel was fitted around the pestle to prevent sample loss through ejection while grinding. Liquid nitrogen was added as needed to mortar to prevent thawing of the tissue.
Mechanical cryogrinding (GG)
Diced cob samples (1.5 g) were ground simultaneously in cycles of 2 min at 200 strokes/min using Spex Sample Prep Geno/Grinder 2000 (SPEX, Metuchen, NJ). Each sample was placed in a 15 ml ball-grinder tube containing three 9.525 mm stainless steel grinding balls (OPS Diagnostics, Lebanon, NJ) that were treated by the manufacturer to remove residual oils and contaminants. The tubes, balls, and stainless steel six-tube rack were pre-chilled in liquid nitrogen before sample insertion. Sample tubes and steel tube rack were kept in a nitrogen bath for 5 min before and after each grinding cycle to reduce tissue thawing. A total of ten cycles were performed, at which point most of the tissue had become a fine powder. Increasing the number of cycles did not improve pulverization any further.
Adapted focused acoustics (AFA)
According to manufacturer protocol for solid samples, diced cob tissue (1.5 g) was inserted into specialized pulverization bags (Cat. No. 520001, Covaris, Woburn, MA), submerged to liquid nitrogen for 1 min, then positioned under an anvil mechanism and hit three times with a mallet, crushing the tissue. The bags were returned to a liquid nitrogen bath for 5 min before being transferred to a specialized AFA vial (Cat. No. 520080, Covaris, Woburn, MA). The sample vial was then placed in a Covaris S220 acoustic focused-ultrasonicator (Covaris, Woburn, MA) that subjected the tissue to 1,000 cycles per burst of 500 W ultrasonic power at a duty factor of 20 over a 1 min period at 4°C to further disrupt the cellular structure of the tissue. The given setup represents the most stringent one allowed by the instrument.
Chemical elution of metabolites
For all three grinding methods, once the tissue sample had become a fine powder, it was suspended in 4.5 ml of methanol with 0.125% formic acid (v/v) and processed according to Kim and Verpoorte (2010). The suspension was vortexed and then placed in a Cole-Parmer ultra-sonicator (Cole-Parmer, Vernon Hills, IL) bath at 40 kHz for 10 min. Suspensions were centrifuged at 21,130 x g for 50 min at 4°C. The supernatant of 3 ml was removed without disturbing the pellet and filtered through a 0.2-micron polytetrafluoroethylene (PTFE) disk syringe filter, and the pellet was discarded. Post filtration, the samples were centrifuged at 21,130 x g for 15 min at 4°C to ensure no precipitate remained. The supernatant was transferred to 1.5 ml tubes in aliquots (150 ml) while being speed-vac dried at room temperature. Blank replicates (extraction buffer only) were subjected to the same procedure as the tissue containing samples.
Liquid chromatography–mass spectrometry analysis (LC-MS)
All samples were resuspended in 50 ml of 2% acetonitrile with 0.1% formic acid (v/v). They were analyzed by an Ultimate 3000 HPLC system directly linked to a LTQ Orbitrap Velos mass spectrometer (both ThermoFisher Scientific, Waltham, MA). Seven ml of each sample were loaded on C18 reversed-phase column, and metabolites were separated via constant flow (0.33 µl.min-1), 60-min long linear gradient of acetonitrile in 0.3% formic acid (2% for 5 min, 2 - 75% for 30 min, 95% for 10 min, 2% for 15 min). Analytes were nebulized via nano-electrospray ionization, and mass spectra were collected in full scan, profile, no fragmentation, positive mode, with the mass resolution set to 100,000.
Principal component analysis of metabolite features
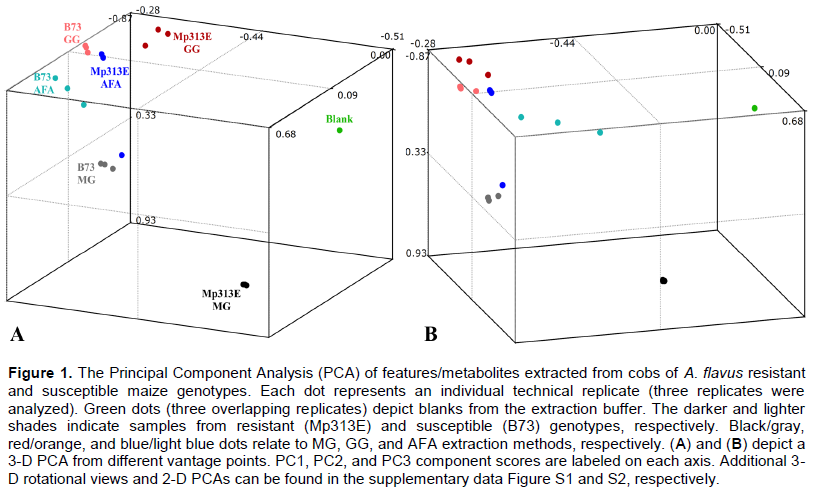
To evaluate methods for extractions of metabolites, it is not necessary to actually identify the compounds. The immediate subjects of following analyses are “features” and their intensities as representative parameters of pertinent metabolites entities. The term “feature” describes an LC-MS signal consisting of retention time (RT) and mass to charge ratio (m/z) (Tautenhahn et al., 2008). Raw LC-MS data files were analyzed by Sieve software (ThermoFisher Scientific, Waltham, MA) using the “Small molecule chromatographic alignment” module capable of performing principal component analysis (PCA) (Figure 1). Critical processing parameters were set as follows: m/z interval 85-900, RT interval 10-50 min., peak time width of 2 min., m/z width 7 ppm, the maximum number of frames 3,000, and alignment minimal intensity 1000.
The PCA analysis results show that grouping of detected features was method-dependent, as demonstrated by the clustering of replicates for each extraction technique, rather than clustering based on genotypes. As indicated by the tight clustering of the manual ground (MG) Mp313E (black dots) and MG B73 replicates (gray dots), manual grinding showed the highest reproducibility among tested methods. The second-best reproducibility was achieved by the mechanical grinding method (GG), where both GG Mp313E (red dots) and GG B73 (orange dots) exhibited close clustering of replicates. Most loosely grouped replicates were the Adaptive Focused Acoustics (AFA) Mp313E (blue dots) and B73 (light blue dots), indicating that the Adaptive Focused Acoustics method of homogenization is the least reproducible. The three Blank replicates (green dots) overlap and appear as a single dot in PCA from any angle, affirming high reproducibility of LC-MS data collection. Therefore, observed differences among the replicates across the methods can be confidently attributed to true differences in extracted metabolomes, rather than to irreproducible LC-MS measurements.
Venn diagrams of unique and overlapping metabolite features
The features detected by Sieve software for each method and genotype were exported to Venny online software software (BioinfoGP Madrid, Spain) (Oliveros, 2007-2015)to create strict Venn diagrams (Figure 2). The number of features detected in Mp313E samples obtained via AFA, GG, and MG method was 1,102; 1,019; and 1,163 respectively, for a total of 1,362. In B73 genotype, respective numbers were 1,110; 926; and 1163, totaling 1,275 features. There was 54.6% and 72.7% overlap among all three methods for Mp313E and B73 genotype, respectively. The manual grinding produced the highest number of method-unique features (8.4% and 9.3% of the total features detected in Mp313E and B73, respectively). The differences among methods in regard to total and unique features are quite small (~ 21% and 9%, respectively, in most extreme cases), but nevertheless, the results point to MG as the most efficient method.
Interestingly, the most primitive method of tissue homogenization, manual grinding (MG), has proven to be the most reproducible and efficient, out of the three tested techniques for maize cob homogenization and single solvent metabolite extraction. It is possible that MG technique dominates “because of”, and not “despite” being a purely manual, human-controlled method. Unlike the “robotic” methods, MG allows for intelligent intervention for the finest grinding via the ability to see and target the remaining larger pieces of cob tissue to achieve a more homogeneous powder sample. Contrary to expectation, mechanical grinding (GG) commonly left small pieces (1 mm in diameter) of intact cob tissue in the ball-grinder tube, regardless of a prolonged time of processing or increasing the number of balls. Similarly, the temperature can be controlled more consistently during manual grinding, as spontaneous additions of liquid nitrogen in response to thawing. During mechanical grinding, nitrogen cooling of utilized hardware occurred before and after each of the pulverization cycles. Local heating due to friction between balls and tissue could have occurred to the level promoting random molecule degradation, affecting the reproducibility and efficiency of the method. The Adapted Focused Acoustics (AFA) method yields were slightly better than for mechanical grinding. Obviously, the sonic bursts are powerful enough to release small molecules from hard cob tissue. However, the reproducibility was lowest, possibly due to random movement of the cob particles within sonication vials, and due to uncontrolled metabolites reactions taking place at a comparatively high temperature (4°C) during tissue disruption.
This study explored a critical initial step in a larger scope metabolomics experiment – single solvent extraction of metabolites from a tissue of interest, particularly the maize cob, which has not been scrutinized yet for its role in molecular defense against pathogens. Three grinding techniques were examined and quantified in regard to their reproducibility, efficiency, and general feature detection for later LC-MSn experiments. Out of those three, the best performing extraction method for future cob metabolomics experiments was ascertained, however careful results validation will be needed to distinguish between true biological phenomenon and artifacts rising from a methodology bias. Future cob metabolite extractions will combine manual and mechanical grinding aspects to maximize feature extraction, reproducibility, and physical effort as the number of samples scales up.
The authors have not declared any conflict of interests.
We thank Dr. Olga Pechanova for her help and guidance in early stages of the project. This work was supported by Genomics of Agricultural Species and their Pest and Pathogens (USDA Award No. 58-6066-6-04) and the National Institute of Food and Agriculture’s Agriculture and Food Research Initiative for Education and Literacy Initiative (Award No. 2017-67011-26081). Additionally, the USDA-ARS Corn Host Plant Resistance Research Unit at Mississippi State University supported field work and sample collection.
REFERENCES
|
Alfaro Y (2000). Response of resistant and susceptible maize genotypes to inoculation with transformed Aspergillus flavus isolates.
|
|
|
|
de Souza LP, Alseekh S, Naake T, Fernie A (2019). Mass Spectrometryâ€Based Untargeted Plant Metabolomics. Current Protocols in Plant Biology 4(4):e20100.
Crossref
|
|
|
|
|
Jones WP, Kinghorn AD (2006). Extraction of plant secondary metabolites. Natural Products Isolation 323-351.
Crossref
|
|
|
|
|
Kim HK, Verpoorte R (2010). Sample preparation for plant metabolomics. Phytochemical Analysis 21(1): 4-13.
Crossref
|
|
|
|
|
Magbanua ZV, Williams WP, Luthe DS (2013). The maize rachis affects Aspergillus flavus spread during ear development. Maydica 58(2):182-188.
|
|
|
|
|
Oliveros JC (2007-2015). An interactive tool for comparing lists with Venn's diagrams.
View
|
|
|
|
|
Pechanova O, Pechan T (2015). Maize-pathogen interactions: an ongoing combat from a proteomics perspective. International Journal of Molecular Sciences. 16(12): 28429-28448.
Crossref
|
|
|
|
|
Pechanova O, Pechan T, Williams WP, Luthe DS (2011). Proteomic analysis of the maize rachis: potential roles of constitutive and induced proteins in resistance to Aspergillus flavus infection and aflatoxin accumulation. Proteomics 11(1): 114-127.
Crossref
|
|
|
|
|
Russell W (1972). Registration of B70 and B73 Parental Lines of Maize1 (Reg. Nos. PL16 and PL17). Crop Science 12(5): 721-721.
Crossref
|
|
|
|
|
Scott GE, Zummo N (1990). Registration of Mp313E parental line of maize. Crop Science 30(6):1378-1378.
Crossref
|
|
|
|
|
Tautenhahn R, Boettcher C, Neumann S (2008). Highly sensitive feature detection for high resolution LC/MS. BMC Bioinformatics 9(1): 504.
Crossref
|
|
|
|
|
Tugizimana F, Steenkamp PA, Piater LA, Dubery IA (2018). Mass spectrometry in untargeted liquid chromatography/mass spectrometry metabolomics: Electrospray ionisation parameters and global coverage of the metabolome. Rapid Communications in Mass Spectrometry 32(2): 121-132.
Crossref
|
|