ABSTRACT
In this study, the genetic diversity and structure of 90 monosporidia mating-type isolates of Sporisorium scitamineum collected from China were comprehensively evaluated based on the DNA extracted from mating-type haploid sporidias using inter-simple sequence repeat (ISSR) polymorphism molecular marker technique. Fifteen selected ISSR primers produced 162 amplified DNA fragments, of which 115 (71.0%) were polymorphic. Polymorphic information content (PIC) values ranged from 0.12 to 0.92 with an average of 0.82, indicating that a moderate degree of genetic diversity exists among these 90 monosporidia mating-type isolates of S. scitamineum collected from China. Genetic differentiation coefficient (Gst) was calculated to be 0.320, indicating that the total genetic variation between populations was 32%, while 68% of the total genetic variation was from within populations. Meanwhile, gene flow (Nm) was 1.063, indicating lower rate of gene flow between populations. Based on the ISSR marker data set, the results of clustering by the unweighted pair group method of arithmetic average and principal component analysis were similar and could divide these 90 S. scitamineum isolates into two main groups. The isolates collected from the same province tend to cluster in the same main groups. This study shows that the genetic differentiation of these 90 monosporidia mating-type isolates of S. scitamineum was highly correlated with the ecological environment of collection sites, and the heterogeneity of ecological environment was the main driving force for the differentiation of S. scitamineum.
Key words: Sugarcane, Sporisorium scitamineum, mating-type haploid sporidia, inter-simple sequence repeat, genetic diversity.
Sugarcane (Saccharum spp. hybrid) is an important economic crop of sugar and ethanol production. China is currently the third largest producer of cane sugar in the world, following Brazil and India. Southwest and southern China, including Guangxi, Yunnan, Guangdong and Hainan provinces, are the major sugarcane-producing areas in China (Chen and Yuan, 2010). Smut disease which is caused by the fungus, Sporisorium scitamineum formerly called Ustilago scitaminea (Stoll et al., 2003), is one of the most intractable and devastating diseases of sugarcane in the world. It causes not only considerable yield loss, but also leads to variety elimination due to susceptibility to the disease (Ferreira and Comstock, 1989). In 1877, it was first reported in Natal during the early days of sugarcane smut in South Africa and now presented in most of the sugarcane-producing areas of the world (Santiago et al., 2009). Only Papua New Guinea is considered as the origin center for Saccharum officinarum and is the main species involved in modern cultivars, and Fiji Islands are still free of this disease (Raboin et al., 2007). Sugarcane smut was first reported in 1932 in Guangzhou, Guangdong province, China (Antoine, 1961). The disease has caused serious problems in sugarcane plantation and sugar production, especially in the last 15 years due to the susceptibility of the sugarcane cultivar ROC22 which occupies more than 50% of the total sugarcane planting area in China. The average of smut infection rate in ROC22 is over 8% in plant cane and 15% in ratoon cane, respectively (Shen et al., 2013; Xu et al., 2014). The disease has now caused the most serious economic loss of sugarcane in China. It is estimated that the economic losses amounted to 8-10 billion dollars per year in China (Shen et al., 2013).
The most efficient and economic method for disease control, including sugarcane smut, is the use of resistant varieties (Wada, 2003; Shen et al., 2014). The investigation of the genetic diversity of S. scitamineum through the use of DNA molecular fingerprinting techniques is extremely important for a better understanding of the structure of pathogen populations and the interactions among host and pathogen and the environment and as a basis for developing smut-resistant sugarcane varieties. During the past ten years, several studies on genetic diversity of S. scitamineum were conducted in Australia (Braithwaite et al., 2004), France (Raboin et al., 2007) and China (Que et al., 2012; Shen et al., 2012a; Xu et al., 2014). Raboin et al. (2007) analyzed a collection of S. scitamineum populations from 15 sugarcane-producing countries for polymorphisms at 17 microsatellite loci. The report showed that the genetic diversity was extremely low among the American and African populations, and high among the Asian populations, particularly those from the Philippines. In addition, this report also showed that the American and African populations all belonged to a single lineage which was also found among some Asian populations. Previous studies (Que et al., 2012; Shen et al., 2012a; Xu et al., 2014) on genetic diversity of S. scitamineum collected from China showed that the molecular diversity of S. scitamineum was associated with its geographical origin. Shen et al. (2012a) also suggested that majority of S. scitamineum in sugarcane-producing regions of Southern China may belong to or are genetically similar to race 3.
S. scitamineum is bisexual and dimorphic (Sun et al., 2014). It can produce diploid spores called teliospores. When it germinates, the teliospore undergoes meiosis and gives rise to a septate promycelium bearing four haploid sporidia (basidiospores). Of the four initial haploid sporidia or basidiospores from each teliospore, two are positive mating-type haploid sporidia, and the other two are negative mating-type haploid sporidia. Teliospore and mating-type haploid sporidia have their respective genetic background. In the previous studies (Que et al., 2012; Xu et al., 2014), DNA was extracted from teliospores of S. scitamineum on the basis of genetic evolution of S. scitamineum from Mainland China. Although, DNA was extracted from mating-type haploid sporidia in the report of Shen et al. (2012a), only 35 samples collected from 6 sugarcane-producing areas in three provinces of China were analyzed. Thus, to better understand the genetic diversity of S. scitamineum from China, analysis of more DNA samples extracted from mating-type haploid sporidia of S. scitamineum collected from more regions and more cultivar/genotypes were needed.
Inter-simple sequence repeat (ISSR) (Zietkiewicz et al., 1994) is the main molecular marker for kinship and population studies. The method has advantages of high-efficiency, sharp sensibility, good reliability and easy-detection. Therefore, the ISSR markers have now been widely used for genetic diversity analysis, line identification, genetic map construction, or other studies in various organisms, including animals (Zamani et al., 2011), plants (Luo et al., 2011) and microorganisms (Velez et al., 2016).
In the present study, ISSR molecular markers were used to study 90 S. scitamineum isolates obtained from 19 sugarcane-producing areas of five provinces in China based on DNA extracted from mating-type haploid sporidia of S. scitamineum. The results offer a better understanding of the genetic diversity and population structure of this pathogen, and provide useful information for breeding of smut-resistant sugarcane varieties.
S. scitamineum sample collection and isolation
A total of 90 monosporidial mating-type isolates derived from 90 single-whips (sori) of sugarcane smut were collected from the five primary sugarcane-producing provinces (19 planting districts) in China in 2014, including Guangxi (45 isolates), Yunnan (16 isolates), Guangdong (16 isolates), Hainan (11 isolates) and Jiangxi (2 isolates) (Table 1 and Figure 1). Guangxi mainly belongs to inland and subtropical climate, Yunnan is located in the inland plateau with diverse climate type, mainly including tropical, subtropical and frost climate, Guangdong is mainly coastal subtropical and northern frost climate, and Hainan belongs to the coastal tropical climate. Monosporidial mating-type isolates of S. scitamineum were prepared according to the previous studies of Moosawi-Jorf and Izadi (2007).
DNA extraction and identification
Monosporidial DNA samples were prepared by using cetyl trimethyl ammonium bromide (CTAB) method as described by Shen et al. (2006). The purified DNA was quantified by measuring the absorbance at 260 nm. The purity of DNA was determined by calculating the ratio of absorbance at 260:280 nm. The DNA samples were then diluted to prepare a working solution of 20-30 ng/μl for PCR analysis, then, stored at -20°C for further use. To ensure that all the DNA samples were from S. scitamineum, these DNA samples we amplified by S. scitamineum specific primers SL and SR (Shen et al., 2012b).
ISSR analysis
Fifteen S. scitamineum ISSR primers used in this study (Table 2) were commercially synthesized by the Sangon Biotech Co. Ltd. (Shanghai, China) and rTaq DNA polymerase was purchased from the TaKaRa Biotechnology Co. Ltd. (Dalian, China). The PCR reaction mixture was performed in a final volume of 25 μl containing 1 μl of genomic DNA (about 20 to 30 ng), 0.3 μl of rTaq DNA polymerase (5 U/μl), 2.5 μl of 10 × PCR reaction buffer (with Mg2+), 2 μl of 2.5 mM dNTP mixture and 1 μl of 5 μM ISSR random primer. The final volume was adjusted to 25 μl with 18.2 μl sterile distilled water (Shen et al., 2012a). PCR amplification was performed on a MyCyclerTM thermal cycler (BIO-RAD inc.), with an initial denaturation at 94°C for 5 min, followed by 35 cyclers each of denaturation at 94°C for 30 s, annealing at 50°C for 45 s and extension at 72°C for 2 min, and a final extension of 72°C for 8 min. The amplification products were electrophoresed on a 1.8% agarose gels with 0.5×TBE buffer at 100 V for 1.5 h along with the DL 2000 DNA markers. The separated DNA fragments were stained with 5% GoldView™ and photographed with a Gene Genius Biomaging System. The experiment was repeated for at least once.
Data analysis
Based on the electrophoresis results of amplification products by ISSR-PCR, the samples with DNA band were marked as “1”, whereas those without DNA band were marked as “0” (only the repeatable bands in the DNA electrophoresis analysis were recorded). Jaccard similarity coefficient, unweighted pair group method with arithmetic mean (UPGMA) cluster analyses and principal component analysis (PCA) were conducted by using NTSYS-pc2.10 analytical software (Rohlf, 2000). Polymorphic information content (PIC) was calculated using formulas described by Botstein et al. (1980). Population genetic parameters such as the observed number of alleles (Na), effective number of alleles (Ne), Nei,s gene diversity index (h), Shannon,s information index (I), genetic differentiation coefficient (Gst) and gene flow (Nm) were used to evaluate the genetic diversity within each population by POPGENE1.31 (Yeh et al., 1999).
Isolation and identification of monosporidial mating-type isolates of S. scitamineum
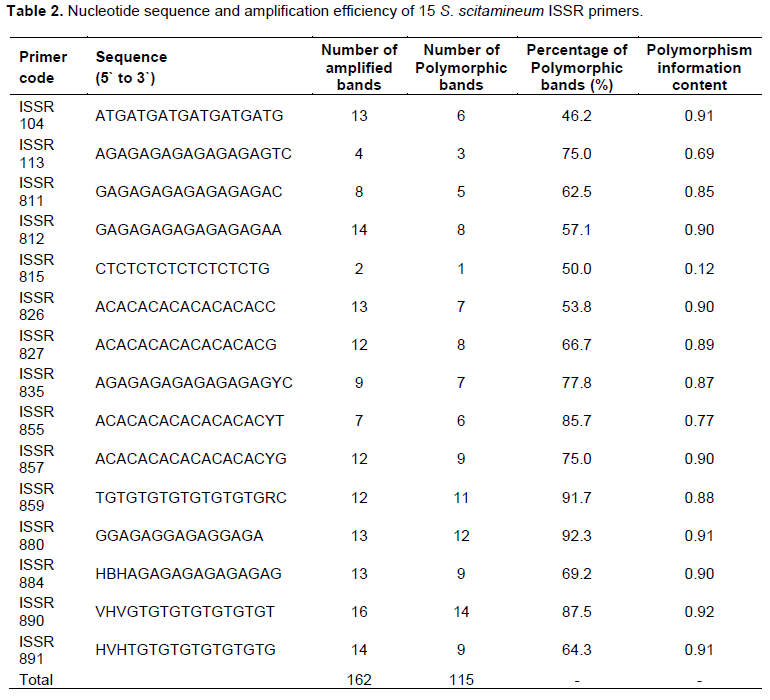
Teliospores began to germinate 20 min after culture. Often, teliospores were germinated on YePS medium after 24 h and produced promycelia and sporidia or produced mycelia. Forty-eight hours after spreading the germinated teliospores on YePS medium, white wool-like monocolonies (dikaryotic mycelium) were produced on the medium (Figure 2a). Yeast-like monosporidial cultures (haploid sporidias) were obtained by streaking of a wool-like monocolony on YePS medium after 48 h at 28°C (Figure 2b). White wool-like dikaryotic mycelium were produced in compatible crosses (that has been postulated to include different mating types), whereas in incompatible crosses (that has been postulated to include the same mating type), the colonies survived as yeast like at the point where two monosporidias cross on YePS medium after 36 h at 28°C (Figure 2c). PCR analysis of the 90 samples by S. scitaminea specific primer pair SL and SR revealed a fragment with expected size of 530 bp, which is present in all the tested samples. Amplified electrophoresis of representative samples is shown in Figure 2d.
ISSR polymorphism analysis in the genetic diversity of mating-type isolates of S. scitamineum
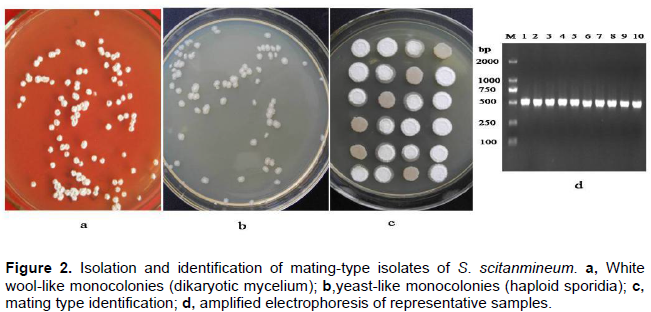
Based on the preliminary results of amplification, 15 ISSR primers (Table 2) were selected for further study of the 80 arbitrarily designed ISSR primers. The selected primers generated clear bands with good polymorphisms in the ISSR analysis of S. scitamineum (90 mating-type isolates). One representative profile (ISSR891) is shown in Figure 3. The number of bands and the degree of polymorphism revealed by each primer are given in Table 2. A total of 162 ISSR bands were generated, of which 115 were polymorphic among the 90 mating-type isolates. Ploymorphic rate of each primer ranged from 46.2 to 92.3%, with an average of 71.0% and the sizes of bands varied from 200 to 3500 bp. The number of ISSR bands generated by each primer was in the range of 2 (ISSR815) to 16 (ISSR890) with an average of 10.8. Among the primers used in this study, the primer ISSR890 generated most bands with 14 polymorphic bands, while the primers ISSR815 and ISSR113 produced only 1 and 3 polymorphic bands, respectively. Overall, the PIC value of these 15 ISSR primers ranged from 0.12 to 0.92 with an average of 0.82. Primer ISSR890 was the most discriminatory with a PIC value of 0.92, whereas ISSR815 had the lowest PIC value of 0.12.
Genetic similarity
Based on the ISSR amplification data, the Jaccard’s genetic similarity coefficients between these 90 S. scitamineum isolates were calculated, and their values varied from 0.52 to 0.98 with an average of 0.75. The genetic similarity coefficients among the vast majority of isolates ranged from 0.76 to 0.95. In general, the genetic similarity among these isolates was higher, while the genetic diversity was moderate. The most genetic similarity (0.98) was found in the isolates number 39 and 40, both of which were isolated from different plants of the same sugarcane cultivar ROC22 at the same geographic location (Tianyang county of Guangxi province), whereas the most dissimilarity (0.52) was observed between the isolates numbers 69 and 87, both of which were isolated from sugarcane cultivar ROC22 at different geographic locations, the isolate number 69 was collected from Liucheng county of Guangxi province (located in North Guangxi, belonging to low altitude frost region) (Figure 1 and Table 1), while the isolate number 87 was isolated from Honghe county of Yunnan province (located in Southeast Yunnan, belonging to high altitude region) (Figure 1 and Table 1).
UPGMA dendrogram analysis
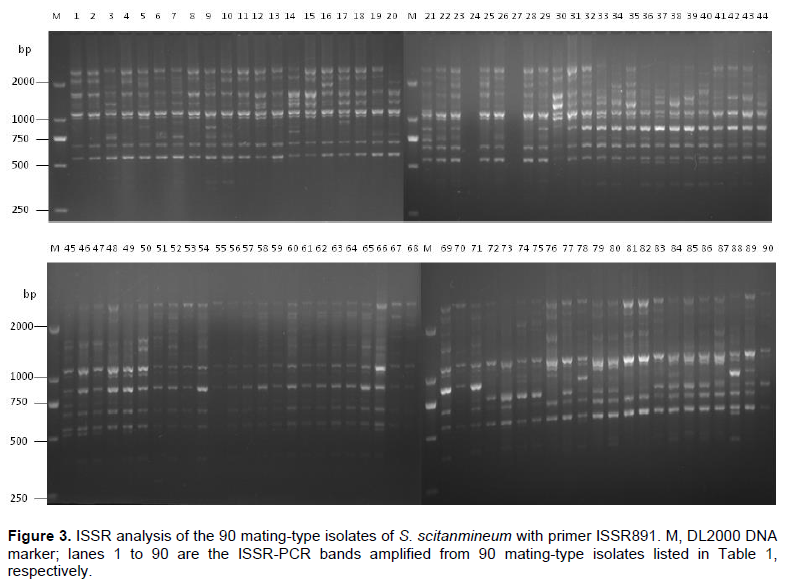
According to the ISSR data set, a dendrogram was generated for these 90 S. scitamineum isolates by UPGMA clustering analysis (Figure 4). The 90 mating-type isolates of S. scitamineum were divided into two main groups (G1 and G2) based on the Jaccard`s similarity coefficient of 0.69 (Figure 4). Among these 90 isolates, 46 (51.1%) were grouped in group 1 (G1), which includes all 10 isolates collected from Leizhou, Guangdong, all 8 isolates from Hepu, Guangxi, and most of the isolates from Hainan (eight of all 11 isolates) and Yunnan (12 of all 16 isolates), together with 6 isolates from Guangxi (3 isolates from Yizhou, 2 isolates from Liucheng and 1 from Longan) and 2 from Wengyuan county of Guangdong province, while 44 isolates (48.9%) were grouped into group 2 (G2), including most Guangxi isolates (31 of all 45 isolates), a small number of isolates from Guangdong (4 isolates), Yunnan (4 isolates) and Hainan (3 isolates), together with all two isolates collected from Jiangxi. It was also observed that isolates from the same sugarcane-growing areas are usually clustered together, for example, isolates numbers 36 to 44 collected from Tianyang county of Guangxi province, isolates numbers 1 to 7 collected from Leizhou county of Guangdong province, isolates numbers 61 to 65 collected from Nanning, Guangxi, and so on (Figure 4).
Principal component analysis
Principal component analysis (PCA) of these 90 S. scitamineum isolates was performed based on ISSR data set (Figure 5). Amongst these 90 isolates, 87 were grouped into two groups (A and B), except for three isolates, numbers 13, 14 and 16, which were collected from Wengyuan county of Guangdong province. The group A contained 45 isolates, which are 11 from Guangdong, 8 from Hainan, 12 from Yunnan and 14 from Guangxi. Group B was composed of 42 isolates, of which 31 are from Guangxi, 4 from Yunnan, 2 from Jiangxi, 3 from Hainan and 2 from Guangdong. As compared to the results of UPGMA analysis, all 45 isolates in the group A were also included in the cluster G1 of UPGMA, while 41 out of 42 in the group B were also included in the cluster G2 of UPGMA, indicating that the results of PCA plot was similar to the results of UPGMA dendrogram.
Genetic diversity and structure analysis
Population genetic parameters (Na, Ne, h, I, pl and r) were conducted to further understand the genetic diversity of these 90 S. scitamineum isolates collected from China (Table 3). The Na value ranged from 1.043 (for Jiangxi population) to 1.673 (for Guangxi population), Ne value varied from 1.031 (for Jiangxi population) to 1.453 (for Guangxi population), h value ranged from 0.018 (for Jiangxi population) to 0.256 (for Guangxi population), I value varied from 0.026 (for Jiangxi population) to 0.377 (for Guangxi population), pl value ranged from 7 (for Jiangxi population) to 109 (for Guangxi population), and r value varied from 4.32 (for Jiangxi population) to 67.28% (for Guangxi population). In addition, the Na, pl and r value were all higher in the population of Yunnan (Na = 1.611, pl = 99 and r = 61.11%) than in the population of Guangdong (Na = 1.593, pl = 96 and r = 59.26%) and Hainan (Na = 1.556, pl = 90 and r = 55.56%), respectively. This indicated that there was higher genetic differentiation in Guangxi and Yunnan populations, moderate genetic diversity in Guangdong and Hainan populations, and lowest genetic diversity in Jiangxi population.
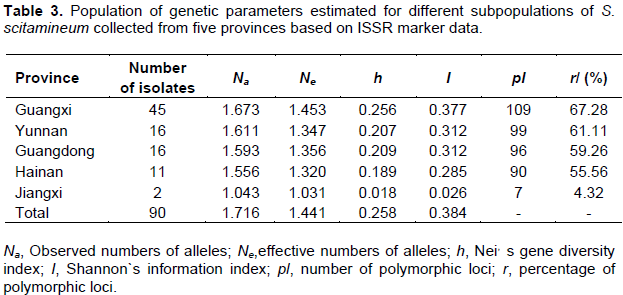
According to the ISSR data set, the total gene diversity (Ht) and gene diversities between subpopulations (Hs) were calculated to be 0.258 and 0.176, respectively. Genetic differentiation coefficient (Gst) was calculated to be 0.320, indicating that only 32% of the total genetic variation originated between populations, while 68% of the total genetic variation was from within populations (Hartl and Clark, 2007). Meanwhile, gene flow (Nm) was calculated to be 1.063, indicating that lower rate of gene flow occurred among the populations (Slatkin, 1985; Qu et al., 2004).
The ISSR molecular marker technique has been proved to be an efficient and economical way to provide molecular data to assess genetic diversity, and it has been used successfully used to determine the genetic relationships for fungus (Velez et al., 2016), plants (Luo et al., 2011) and animals (Zamani et al., 2011). In the present study, the ISSR molecular marker was first comprehensively applied to assess the genetic diversity of mating-type isolates of S. scitanmineum. It was found that the rate of polymorphic bands generated among 90 mating-type isolates of S. scitanmineum collected from China was 71.0%. It was relatively lower than that of previous report (86.8% polymorphism) by Xu et al. (2014) which used ISSR to assess the genetic variation of 100 teliospore isolates of S. scitanmineum from Mainland China. This is in agreement with the results of Shen et al. (2012a) study (72.7% polymorphism) which used ISSR to assess the genetic diversity based on 35 mating-type isolates of S. scitanmineum from Southern China. The difference in polymorphic rate may be due to the fact that Xu et al. (2014) used template DNA extracted from teliospores (diploid spores) of S. scitanmineum for ISSR amplification, while in the current study, DNA samples were from a single mating-type haploid of S. scitanmineum. It was speculated that there was richer genetic variation in dikaryotic teliospores than in monosporidial mating-type isolates of S. scitanmineum. The fact showed that further studies on genetic diversity of monosporidial mating-type isolates of S. scitanmineum are absolutely necessary.
The results of UPGMA and PCA analysis from this study showed that the mating-type isolates of S. scitamineum collected from the same sugarcane-growing areas are usually clustered together. Thirty-five mating-type isolates of S. scitamineum were isolated from the same sugarcane cultivar ROC22 (host) but were planted in 12 different sugarcane producing-areas, including Guangxi, Yunnan, Guangdong and Hainan. These were almost not clustered into the same group, but also tended to cluster according to their geographical origin which indicated that the molecular genetic diversity of the 90 mating-type isolates of S. scitanmineum collected from China was associated with their geographical origins at a large degree and almost not with host association, which was basically consistent with the conclusions of previous studies based on template DNA extracted from dikaryotic teliospore isolates of S. Scitamineum collected from Mainland China (Que et al., 2012; Xu et al., 2014). The mating-type isolates collected from the same or similar ecological environment can always be clustered together. For example, isolates collected from Hepu County, Guangxi Province and isolates collected from Leizhou County, Guangdong Province were always clustered together, although these two counties belong to different provinces, however, these two places are relatively close, with similar ecological environment, which belongs to the low latitude hills, tropical arid climate. In addition, the isolates obtained from Wenyuan County, Guangdong Province and the isolates from Leizhou County, Guangdong Province cannot be clustered together. Although, the two counties belong to Guangdong Province, but the geographical distance between the two places is far away and their ecological environment is different. Leizhou is located in the southwest of Guangdong Province, which belongs to the tropical marine climate, while Wengyuan is located in the northern mountainous area of Guangdong Province, which belongs to the northern margin of the frost area. Thus, it is deduced that there is a high correlation between the genetic differentiation of S. scitanmineum and the ecological environment, and the heterogeneity of the ecological environment is the main motivation of the differentiation of S. scitanmineum.
Population genetic analysis, which can provide strong evidence about genetic diversity at the population or subpopulation levels, should be helpful for the understanding of the genetic structure of the studied populations (Raboin et al., 2007; Xu et al., 2014). Population genetic parameters, (Na, Ne, h and I), reflected the diversity and differentiation within and between the populations; the higher the index, the greater the genetic diversity. According to corresponding parameter values from this study, it is found that mating-type isolates from Guangxi, Yunnan and Guangdong provinces have higher genetic diversity than those from Hainan and Jiangxi provinces, which was similar to the results of a previous study by Xu et al. (2014) based on teliospore isolates of S. scitanmineum collected from China. Previous reports concluded that a gene flow index of Nm>1 would be indicative of no significant differentiation among populations (Slatkin, 1985; Whitlock and Mccauley, 1999). The gene flow index in this study was moderate (Nm=1.063), indicating a high level of genetic diversity within populations that was not prone to the genetic drift. This was further confirmed by the low level of interpopulation genetic differentiation manifested by the low gene differentiation coefficient (Gst) among populations (0.320). Therefore, it is deduced that the genetic diversity among the 90 mating-type isolates of S. scitanmineum had existed mainly within the populations.
Therefore a high correlation exits between the genetic differentiation of S. scitanmineum and their geographical origin in China, which is mainly derived from internal populations. Thus, it is suggested that S. scitanmineum quarantine is necessary in the future when sugarcane varieties or germplasm should be introduced or exchanged from different provinces of China or different ecological regions of the same province to prevent the introduction of new genetic groups of S. scitanmineum with the introduction of sugarcane varieties, because of different genetic groups of S. scitanmineum may exist pathogenicity differentiation or new physiological races.
In summary, the study confirms that there exists a high genetic differentiation among 90 monosporidia mating-type isolates of S. scitamineum from China within populations. The genetic differentiation is highly correlated with the ecological environment of collection sites of isolates, and the heterogeneity of ecological environment is the main driving force for differentiation of S. scitamineum. The present study provided useful genetic information about S. scitamineum isolates from China, which is useful for developing smut-resistant sugarcane breeding and guiding the release of sugarcane varieties.
The authors have not declared any conflict of interests.
REFERENCES
|
Antoine R (1961). Smut. In. Martin JP, Abbott EV, Hughes CG (eds) Sugarcane Diseases of the World. Elsevier Press, Amsterdam, Netherlands. pp. 327-354.
|
|
|
|
Botstein D, White RI, Skolnick M, Davis RW (1980). Construction of a genetic linkage map in man using restriction fragment length polymorphisms. Am. J. Hum. Genet. 32:314-331.
|
|
|
|
|
Braithwaite KS, Bakkeren G, Croft BJ, Brumbley SM (2004). Genetic variation in a worldwide collection of the sugarcane smut fungus Ustilago scitaminea. Proc. Aust. Soc. Sugar Cane Technol. 26:1-9.
|
|
|
|
|
Chen RK, Yuan ZN (2010). Sugarcane production and research in China. Int. Sugar J. 112:452-457.
|
|
|
|
|
Ferreira SA, Comstockand JC (1989). Smut. In. Ricaud C, Egan BT, Gillaspie Jr AG, Hughes CG (eds) Disease of Sugarcane. Elsevier Press, Amsterdam, Netherlands. pp. 211-229.
Crossref
|
|
|
|
|
Hartl DL, Clark AG (2007). Principles of Population Genetics. Sinauer Associates.
|
|
|
|
|
Luo C, He XH, Chen H, Ou SJ, Gao MP, Brown JS, Tondo CT, Schnell RJ (2011). Genetic diversity of mango cultivars estimated using SCoT and ISSR markers. Biochem. Syst. Ecol. 39:676-684.
Crossref
|
|
|
|
|
Moosawi-Jorf SA, Izadi MB (2007). In vitro detection of yeast-like and mycelial colonies of Ustilago scitaminea in tissue-cultured plantlets of sugarcane using polymerase chain reaction. J. Appl. Sci. 7:3768-3773.
Crossref
|
|
|
|
|
Qu RZ, Hou L, LÇš HL, Li HY (2004). The gene flow of population genetic structure. Hereditas 26:377-382.
|
|
|
|
|
Que YX, Xu LP, Lin JW, Chen RK, Grisham MP (2012). Molecular variation of Sporisorium scitamineum in Mainland China revealed by RAPD and SRAP markers. Plant Dis. 96:1519-1525.
Crossref
|
|
|
|
|
Raboin LM, Selvi A, Oliveira KM, Paulet F, Calatayud C, Zapater MF, Brottier P, Luzaram R, Garsmeur O, Carlier J, D`Hont A (2007). Evidence for the dispersal of a unique lineage from Asia to America and Africa in the sugarcane fungal pathogen Ustilago scitaminea. Fungal Genet. Biol. 44:64-76.
Crossref
|
|
|
|
|
Rohlf FJ (2000). NTSYS-pc: Numercial Taxonomy and Multivariate Analysis System.Version 2.1. Exeter Publications, New York, USA.
|
|
|
|
|
Santiago R, Armas R, Fontaniella B, Vicente C, Legaz ME (2009). Changes in soluble and cell wall-bound hydroxycinnamic and hydroxybenzoic acids in sugarcane cultivars inoculated with Sporisorium scitamineum sporidia. Euro. J. Plant Pathol. 124:139-450.
Crossref
|
|
|
|
|
Shen WK, Deng HH, Li QW, Yang ZD, Jiang ZD (2014). Evaluation of BC1 and BC2 from the crossing Erianthus arundinaceus with Saccharum for resistance to sugarcane smut caused by Sporisorium scitamineum. Trop. Plant Pathol. 39:368-373.
Crossref
|
|
|
|
|
Shen WK, Jiang ZD, Deng HH, Liu R (2013). Research progress on sugarcane smut disease and Sporisorium scitamineum. Chin. J. Trop. Crops 34:2063-2068.
|
|
|
|
|
Shen WK, Xi PG, Li MH, Liu R, Sun LH, Jiang ZD, Zhang LH (2012a). Genetic diversity of Sporisorium scitamineum in Southern China revealed by combined ISSR and RAPD analysis. Afr. J. Biotechnol. 11:11693-11703.
|
|
|
|
|
Shen WK, Xi PG, Li MH, Sun LH, Zhang LH, Jiang ZD (2012b). Development of a sensitive nested-PCR assay for detection of Sporisorium scitamineum. Afr. J. Biotechnol. 11:10541-10547.
|
|
|
|
|
Shen WK, Zhou GH, Deng HH, Zhou LY (2006). Detection of sugarcane ratoon stunting disease pathogen with PCR and nucleotide sequence analysis. Chin. Agric. Sci. Bull. 22:413-416.
|
|
|
|
|
Slatkin M (1985). Gene flow in natural populations. Annu. Rev. Ecol. Evol. Syst. 16:393-430.
Crossref
|
|
|
|
|
Stoll M, Piepenbring M, Begerow D, Oberwinkler F (2003). Molecular phylogeny of Ustilago and Sporisorium species (Basidiomycota, Ustilaginales) based on internal transcribed spacer (ITS) sequences. Can. J. Bot. 81:976-984.
Crossref
|
|
|
|
|
Sun LH, Yan MX, Ding ZJ, Liu YB, Du MG, Xi PG, Liao JL, Ji LH, Jiang ZD (2014). Improved dominant selection markers and co-culturing conditions for efficient Agrobacterium tumefaciens-mediated transformation of Sporisorium scitamineum. Biotechnol. Lett. 36:1309-1314.
Crossref
|
|
|
|
|
Velez P, Quintero CA, Merino G, Pineda JG, Gonzalez MC (2016). An ISSR-based approach to assess genetic diversity in the marine arenicolous fungus Corollospora maritima sensu lato. Mycoscience 57:187-195.
Crossref
|
|
|
|
|
Wada AC (2003). Control of sugarcane smut disease in Nigeria with fungicides. Crop Prot. 22:45-49.
Crossref
|
|
|
|
|
Whitlock MC, Mccauley DE (1999). Indirect measures of gene flow and migration: FST not equal to 1/ (4Nm + 1). Heredity 82:117-125.
Crossref
|
|
|
|
|
Xu LP, Lu YH, You Q, Liu XL, Grisham MP, Pan YB, Que YX (2014). Biogeographical variation and population genetic structure of Sporisorium scitamineum in Mainland China: insights from ISSR and SP-SRAP markers. Sci. World J. Article ID 296020.
|
|
|
|
|
Yeh FC, Yang RC, Boyle TBJ, Ye ZH, Mao JX (1999). POPGENE, version 1.31. The user friendly shareware for population genetic analysis. University of Alberta and Centre for International Forestry Research 28:45-47.
|
|
|
|
|
Zamani P, Akhondi M, Mohammadabadi MR, Saki AA, Ershadi A, Banabazi MH, Abdolmohammadi AR (2011). Genetic variation of Mehraban sheep using two intersimple sequence repeat (ISSR) markers. Afr. J. Biotechnol. 10:1812-1817.
|
|
|
|
|
Zietkiewicz E, Rafalski A, Labuda D (1994). Genome fingerprinting by simple sequence repeat (SSR) anchored polymerize chain reaction amplification. Genomics 20:176-183.
Crossref
|
|