ABSTRACT
The present work aims to identify common bean accessions resistant to anthracnose by incompatibility reaction evaluations to races 73 and 2047 of C. lindemuthianum, associated with using of SF101072 and SAS13950 molecular markers linked, respectively, to gene Co-10 and allele Co-42. A total of 75 common bean accessions from Common Bean Germplasm Bank (CBGB) of Núcleo de Pesquisa Aplicada à Agricultura (Nupagri) were evaluated. The results showed that 14 accessions were resistant to both races 73 and 2047 of C. lindemuthianum, 36 exhibited resistances only to race 2047, and 25 revealed incompatibility reaction to race 73. Joint analysis between incompatibility phenotypic reactions and the presence of markers allowed identifying 13 resistant accessions to races 73 and 21 to race 2047, indicating the occurrence of Co-10 (renamed Co-34) and Co-42, respectively. A total of 32 accessions were resistant without the presence of markers. Among the 75 common bean accessions evaluated, important sources of resistance to C. lindemuthianum, from Andean and Mesoamerican regions, were identified and have promising potential in breeding programs.
Key words: Phaseolus vulgaris, anthracnose, molecular marker, sequence characterized amplified regions for amplification of specific band (SCAR).
Anthracnose is one of the most important common bean diseases (Phaseolus vulgaris L.) worldwide, causing crop yield loss up to 100% in regions with prevailing high humidity and moderate temperatures (13 to 27°C) (Vieira and Paula Júnior, 2004; Mendéz-Vigo et al., 2005). This disease is usually problematic in tropical and subtropical regions, where common bean is used as main protein source for human feed (Mendéz-Vigo et al., 2005), being considered endemic in Africa, Asia, Australia, and some other Latin American countries, such as Brazil (Vieira and Paula Júnior, 2004; Mahuku and Riascos, 2004).
Colletotrichum lindemuthianum (Sacc. & Magnus) Scrib. the causal agent of anthracnose in common bean, is an imperfect fungus characterized by its high pathogenic variability, which has approximately 120 races identified in the world (Mahuku and Riascos, 2004; Hernández-Godinez et al., 1998; Ferreira et al., 2008). In Brazil, it has already been identified in more than 50 races of C. lindemuthianum (Alzate-Marin and Sartorato, 2004; Damasceno and Silva et al., 2007; Gonçalves-Vidigal et al., 2008a; Sansigolo et al., 2008), considering that 64, 65, 73, 81, 87 and 89 pathotypes are the most frequent ones, mainly found in States of Paraná, Santa Catarina, Goiás and Distrito Federal.
The high pathogenic variability revealed by C. lindemuthianum, which has not been completely elucidated, is attributed to its parasexual cycle (Rodríguez-Guerra et al., 2004). The pathogen’s reproduction mechanism is responsible for continuous resistance enhancement, mainly observed in commercial cultivars, since most of them present monogenic resistance that is easily overcame by emerging races (Rodríguez-Guerra et al., 2003). However, among the diverse strategies adopted to control anthracnose, genetic resistance is considered the most efficient and economical (Mahuku et al., 2002). For that, common bean genetic breeding programs in Brazil and worldwide, through the effort of many researchers, have been constantly seeking for new sources of genetic resistance to this pathogen.
Previous studies have identified 19 dominant anthracnose resistance genes. Some of these genes were found on Mesoamerican beans while other genes were discovered on Andean beans. The Mesoamerican genes include: Co-2, Co-3 (and its alleles Co-32, Co-33, Co-34, Co-35), Co-4 (and its alleles Co-42, Co-43), Co-5 (and its allele Co-52), Co-6, Co-10 (renamed as Co-34; Gonçalves-Vidigal et al., 2013), Co-11, Co-15, Co-16, Co-17, Co-u, and Co-v and the Andean genes Co-1 (and its alleles Co-12, Co-13, Co-14, Co-15), Co-12, Co-13, Co-14, Co-15, Co-x, Co-w, Co-y, and Co-z (Geffroy, 1997; Geffroy et al., 1999, 2008; Kelly and Vallejo, 2004; Gonçalves-Vidigal and Kelly, 2006; Gonçalves-Vidigal et al., 2007, 2008b, 2009, 2011, 2013; Sousa et al., 2014, 2015; Trabanco et al., 2015; Coimbra-Gonçalves et al., 2016). Among these genes, Co-10 (renamed as Co-34) possesses ample resistance spectrum and is considered incompatible to 17 races of C. lindemuthianum normally found in Brazil regions, especially race 73 (Alzate-Marin et al., 2003; Costa et al., 2003).
On the other hand, allele Co-42 is considered the most effective to control this disease, showing a resistance index of 97% when tested with 34 races of C. lindemuthianum (Balardin and Kelly, 1998), pointing out of them race 2047. This race was firstly described by Mahuku and Riascos (2004), in isolates from Costa Rica, and it is considered one of the most virulent ones because it is capable of overcoming the anthracnose’s resistance conferred by seven genes (Co-1, Co-2, Co-3, Co-4, Co-5, Co-6 and Co-11) and five alleles (Co-12, Co-13, Co-15, Co-33 e Co-43).
In addition to gene identification by conventional methods, the use of molecular markers to characterize resistant genes to anthracnose has significantly contributed for initial steps of breeding programs by reducing the time and costs involved during the whole process. This occurs because DNA markers are closed linked to genes and are not influenced by environmental factors and they show epistatic or minimum/none pleitropic effects (Agarwal et al., 2008).
Among the available molecular markers, the ones denominated as SCAR (Sequence characterized amplified regions for amplification of specific band) have been playing great importance on common bean analyses. Until now, there are 14 SCAR markers linked to anthracnose resistance genes, which are: SEACT/MCCA, SCAreoli1000, SQ41440, SW12700, SY20830, SC08910, SAS13950, SH181100, SBB141150/1050, SAB3400, SZ20845, SZ04567, SB12350 and SF101072 21. SCAR markers have been optimized in breeding programs that search for anthracnose resistant cultivars by implanting assisted backcrosses programs (Miklas and Kelly, 2002), during characterization of accessions in the beginning of the selection process or to obtain superior lineages (Beraldo et al., 2009).
Therefore, the present work’s objective was to identify common bean accessions resistant to anthracnose through evaluations of phenotypic reactions to races 73 and 2047 of C. lindemuthianum associated to the using of SF101072 and SAS13950 molecular markers linked to Co-10 and Co-42, respectively.
Plant material
Identification of resistance sources to races 73 and 2047 of C. lindemuthianum was carried out by evaluation of 75 common bean accessions from CBGB of Nupagri, Universidade Estadual de Maringá (Table 1).
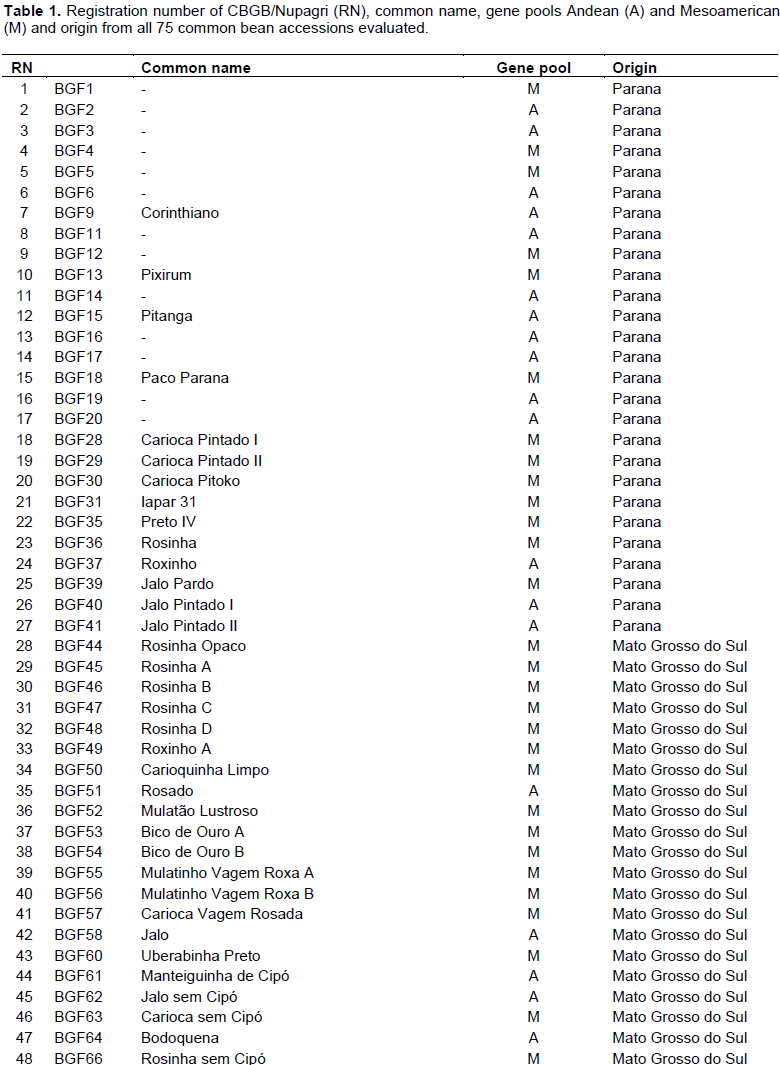

Among the 75 studied accessions, 27 were collected in Mato Grosso do Sul State and kindly provided by Empresa Brasileira de Pesquisa Agropecuária (Embrapa), Centro Nacional de Pesquisa de Arroz e Feijão (CNPAF) according to Material Transference Deal – II (MTD-II). A total of 18 accessions from active germplasm bank were given by Empresa de Pesquisa Agropecuária e Extensão Rural de Santa Catarina (Epagri), which were collected in Santa Catarina State. The accessions BGF1 to BGF 20 were collected in the region of Toledo in Paraná State by the agronomist Rodrigo Garcia and donated to the active common bean germplasm bank of Nupagri, whereas the other 13 accessions used in this study already belong to Nupagri’s germplasm bank.
Seeds from each accession were sown in polyethylene vases (48 × 30 × 11 cm) containing a mixture of previously fertilized and sterilized substrate. The plant vases were kept in a greenhouse until the first trifoliolate leaves (stage V3) were fully expanded. At that time, the plants were inoculated with race 73 and 2047 of C. lindemuthianum and this procedure was repeated for two times. Seedlings were grown under natural light in greenhouses supplemented by 400 w high-pressure sodium lamps giving total light intensity of 115 μmoles m-2 s-1 for 7 to 10 days until they reached the first trifoliate leaf stage. Subsequently, inoculations with races 73 and 2047 of C. lindemuthianum were carried out and analyzed. Finally, we conducted molecular analysis through SCAR SF101072 and SAS13950 markers.
Preparation of samples for DNA extraction
Ten seeds of each accession were grown in plastic trays containing peat moss and vermiculite, and kept in greenhouse until they reached the first trifoliate leaf stage. After this period, a young leaflet of each seedling was individually collected with 1.5 mL plastic micro tubes frozen in liquid nitrogen and stored in freezer (-20°C) for DNA extraction. Subsequently, the trays were transferred to mist chamber with controlled temperature (22 ± 2°C), and later they were inoculated with spores suspension of races 73 and 2047 of C. lindemuthianum.
Inoculum preparation and virulence characterization
Inoculum was prepared according to the methodology proposed by Cárdenas et al. (1964), which consists on multiplying spores of each C. lindemuthianum pathotype in test tubes containing sterilized pods partially immersed in agar-water (AW) culture media. After inoculation, test tubes were incubated for 14 days at 20 ± 2ºC. After incubation period, pods from each test tube were removed using tweezers and transferred to a Beaker containing sterile distilled water. Double gauze was used to filter the suspensions obtained for each pathotype, and the spore suspension was adjusted to 1.2 x 106 spores mL-1 through dilutions with distilled sterile water. Each pathogen was inoculated separately using small brushes (Tigre® model 266, number 14). After inoculation, the plants were placed in a mist chamber for 48 h at a temperature of 20 ± 2°C with light controlled at 12 h of daylight and 12 h of darkness (light intensity of 300 micromoles m-2 s-1 at a height of 1 m) and a relative humidity of 95%. After the incubation period, the inoculated plants were transferred to open-air benches at a temperature of 22°C with artificial light (12 h of daylight at 25°C), where they remained for 3 days before visual symptom assessment. Anthracnose disease reactions were rated visually using a scale from 1 to 9 (Pastor-Corrales et al., 1995). The plants scored from 1 to 3 were considered resistant, whereas the ones scored from as 4 to 9 were susceptible.
Genomic DNA extraction and analysis using SCAR markers
DNA extraction was carried out based on methodology proposed by Afanador et al. (1993). All amplification reactions were performed with a thermal cycler (MJ Research Inc., Waltham, MA). The polymerase chain reaction (PCR) program for SF101072 consisted of 3 min at 94°C, 35 cycles of 15 s at 94°C, 1 min at 65°C, and 90 s at 72°C, followed by a 7 min extension at 72°C and 4 min at 4°C (Corrêa et al., 2000). PCRs were performed in 25 µL total reaction volumes containing 40 ng total DNA; 0.2 mM each dNTP; standard Taq buffer containing 1.5 mM MgCl2 and 0.2 µM forward primer and reverse primer; and 1 unit of Taq DNA polymerase. Following the addition of 2 µL loading buffer (30% glycerol and 0.25% bromophenol blue), The PCR products from SF10 were visualized on agarose gels. Amplification reactions consisted in a total volume of 25 µL each, were conducted for SAS13950 as proposed by Young et al. (1998). The PCR products from SAS13950 were fractionated in agarose gel 1.2% prepared with TAE 1X buffer (40mM Tris-acetate, 20 mM acetic acid and 1mM EDTA), containing ethidium bromide (0.02%). The DNA bands were visualized under ultraviolet light, and digital images were recorded with an L-PIX Image EX model (Loccus Biotecnologia - Loccus do Brasil, Cotia, SP, Brazil). Gel evaluations were conducted to identify accessions with the presence of 1072 bp and 950 bp bands, which correspond to markers SF10 and SAS13, respectively.
Phenotypic data and SCAR markers were analyzed as follow: resistant plants with marker (R+), resistant plant without marker (R-), susceptible plants with marker (S+) and susceptible plant without marker (S-).
Incompatibility reaction of common bean accessions to races 73 and 2047 of C. lindemuthianum
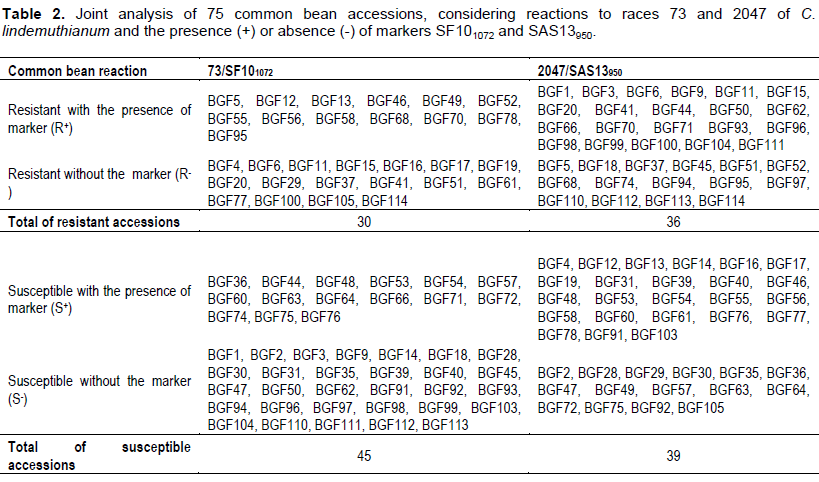
Table 2 shows the phenotypic reactions to races 73 and 2047 of C. lindemuthianum. Based on the results, it was observed that out of the 75 analyzed accessions 30 of them were resistant to race 73, whereas 36 were to race 2047. It was also noted that 14 accessions showed resistance to both races.
Among the 30 accessions resistant to race 73, 15 are Mesoamerican and 15 are Andean. The Andean accessions from Manteigão commercial group prevailed with 12 resistant accessions (BGF 6, BGF 11, BGF 16, BGF 17, BGF 20, Pitanga, Jalo Pintado II, Jalo, Manteiguinha de cipó, Manteiguinha com cipó, Bolinha and Amendoim Cavalo), whereas among Mesoamerican commercial groups Preto (BGF 4, Iapar 44, Crioulo 159, Crioulo Brilhoso Ponte Serrada and FC 117) and Roxinho (BGF 12, Roxinho and Roxinho A) revealed greater importance. The other accessions belong to commercial groups of Rosinha (BGF 5 and Rosinha B, Pixirum and Rosado), Carioca (Carioca Pintado II and Aporé), Mulatinho (Mulatinho Vagem Roxa A and Mulatinho Vagem Roxa B),) and Pardo (Mulatão Lustroso). It is important to mention that 14 accessions are from Paraná, 10 from Mato Grosso do Sul, four from Santa Catarina and two are commercial cultivars (Aporé and Iapar 44).
In the incompatibility reaction to race 2047, the analysis verified that 26 accessions belong to Andean gene pool, while 10 belong to the Mesoamerican. Originally, 14 from these accessions were collected in Santa Catarina, 11 in Parana State and the others in Mato Grosso do Sul. The commercial group with greater number of resistant accessions was Preto (BGF 3, Crioulo Manteiga, Crioulo Brilhoso, Crioulo 159, CF 75, FC 2016, FC 2001, FC 2045, Crioulo Brilhoso Ponte Serrada, Preto Redondo CN694 FC 1212, Azulão Ponte Serrada and Azulão Ab. Luz), from which 11 accessions are from Santa Catarina of the Epagri’s common bean breeding program. The second group with greater number of accessions with resistance was Manteigão (BGF 6, BGF 11, BGF 20, Pitanga, Jalo Pintado II, Jalo sem Cipó, Manteiga com Cipó, Bolinha and Amendoim Cavalo).
An important observation obtained from this study is that out of the 75 analyzed accessions, 14 of them were resistant to both races 73 and 2047 of C. lindemuthianum. Besides that, 10 of them are Andean and belong to commercial group Manteigão (BGF 6, BGF 11, BGF 20, Pitanga, Jalo Pintado II, Manteiga com cipó, Bolinha and Amendoim Cavalo), group Rosinha (Rosado) and group Roxinho (Roxinho). On the other hand, four accessions are Mesoamerican and are allocated in the commercial groups of Preto (Crioulo 159 and Crioulo Brilhoso Ponte Serrada), Rosinha (BGF 5) and Pardo (Mulatão Lustroso).
A total of 75 accessions were analyzed, from which 44 are Mesoamerican and 31 Andean. According to the results obtained, when inoculated with race 73, 42% of Andean accessions and 38.6% of Mesoamerican accessions were resistant. On the other hand, when inoculation was carried out with race 2047, 61% of Andean accessions and 38.6% of Mesoamerican accessions demonstrated incompatibility to this race. It is important to point out that both races 73 and 2047 are Mesoamerican. Melotto and Kelly (2000) have affirmed that Andean accessions tend to be more resistant than Mesoamerican isolates, a fact that was evident when the evaluated accessions were tested with race 2047.
Common bean was independently domesticated from weedy wild beans in two separate geographic centers, the Mesoamerican, extending from Mexico to Colombia; and the Andean, extending from Colombia to Argentina (Gepts and Debouck, 1991). Correspondingly, two genes pools known as the Mesoamerican and the Andean have been described for common bean. Beans from the Mesoamerican gene pool are small to medium-seeded and contain significantly wider genetic diversity than the mostly large-seeded Andean beans (Beebe et al., 2000; 2001; Chacon et al., 2005). Similarly, studies of the virulence and genetic diversity of C. lindemuthianum using differential cultivars and molecular techniques reveal that the diversity of C. lindemuthianum, as well as that of the rust (Uromyces appendiculatus) and angular leaf spot (Phaeoisariopsis griseola) pathogens, segregates into two distinct groups that mirror the diversity of their common bean host (Guzman et al., 1995). These two groups have been also named Andean and Mesoamerican because they correspond to the Andean and Mesoamerican gene pools of their common bean host. Andean races of the anthracnose pathogen are usually isolated from large-seeded beans that belong to the Andean gene pool. Conversely, Mesoamerican races are often, but not always, isolated from small or medium-seeded beans belonging to the Mesoamerican gene pool. Mesoamerican races of this pathogen exhibit considerably greater virulence and genetic diversity than the Andean races. More importantly, Andean races of this pathogen are compatible only or mostly with the Andean beans, while Mesoamerican races are compatible with Mesoamerican and Andean beans, but are generally more virulent on the former. Thus, when breeding beans with anthracnose resistance to Mesoamerican races of C. lindemuthianum, Andean anthracnose resistance loci are extremely valuable. The Andean Co-1 resistance loci have been very valuable in breeding Mesoamerican beans with anthracnose resistance, particularly in production countries where Mesoamerican races of the anthracnose pathogen predominate (Kelly and Vallejo, 2004). Conversely, when controlling Andean races, anthracnose resistance loci from Mesoamerican beans are very important. It has been posited that the Andean and Mesoamerican races of C. lindemuthianum have evolved separately; Andean races with Andean beans in South America and Mesoamerican races with Mesoamerican beans, respectively. The previous discussion highlights the need to identify and characterize additional resistant genes in Andean beans to complement the many anthracnose resistance genes available in Mesoamerican common beans.
Joint analysis between incompatibility reaction to races 73 and 2047 and SCAR molecular markers Joint analysis between incompatibility reaction and the presence of molecular markers SF101072 and SAS13950 are shown in Table 2.
Incompatibility reaction of accessions to race 73 and data regarding SF101072 marker revealed that 17.3% of resistant accessions showed the presence of the marker linked to gene Co-10 (renamed Co-34). Similarly, SAS13950 marker was efficient to identify 28% of accessions resistant to race 2047. The other accessions that exhibited a marker, either linked to gene Co-34 or Co-42 allele, were considered susceptible to races 73 and 2047. In addition, no marker was detected in 22.7 and 20% of accessions considered resistant to races 73 and 2047, respectively.
The molecular analysis revealed that the resistant accessions, in which amplification of marker was noted, possess gene Co-34 and/or Co-42 allele. Therefore, it was observed the occurrence of resistant accessions did not present the molecular marker might had happened due to resistance breaking between gene and marker by recombination process (Melotto et al., 2004). This fact could indicate that these 29 accessions may have different resistance gene(s) from those evaluated in this study.
As noted already by Melotto and Kelly (2001), the fact that susceptible accessions have amplified the markers might have occurred due to intragenic recombination. These authors have demonstrated intragenic recombination in Co-4 locus, which resulted in susceptible recombinants capable to amplify the marker even at a lower rate. The Co-4 locus contains several paralogs of the COK-4 gene that is predicted to code for a receptor-like kinase (RLK). The predicted COK-4 protein is highly similar to FERONIA (FER), a membrane-localized protein of Arabidopsis, as they cluster in a single clade of the RLK phylogenetic tree (Melotto and Kelly, 2001; Melotto et al., 2004; Oblessuc et al., 2015). Receptor-like kinases (RLKs) are important pattern recognition receptor (PRRs) that play a role in self and non-self-recognition, including the perception of hormones (Shiu and Bleecker, 2001), pathogen associated molecular patterns (PAMPs) and pathogen effectors.
Joint analysis between incompatibility reactions to races 73 and 2047 and molecular markers pointed out that many accessions possess at least one Mesoamerican resistance gene, as: Co-4, Co-5, Co-6, Co-10 and Co-42, or another gene not yet identified. Indeed, these accessions will have to be analyzed through allelism tests to determine the independence of the anthracnose resistance gene in these accessions from the known anthracnose resistance genes in common bean. Additionally, wide-genome association using Single Nucleotide Polymorphism (SNP) assay and Kompetitive Allele Specific PCR (KASP) markers will be conducted for genotyping these accessions and mapping resistance genes to C. lindemuthianum.
Common bean accessions analyzed in this study have shown incompatibility to two pathotypes that consist of important anthracnose resistance sources, especially those that present resistance to race 2047. Among the 75 common bean accessions that belong to Nupagri’s Germplasm Bank were identified important resistance sources to C. lindemuthianum of Andean and Mesoamerican origins that could be available for breeding programs. In addition, the identification of resistant accessions without association to analyzed markers points out that they could represent new resistance sources to anthracnose. Therefore, genetic characterization of these accessions is essential for their future using in breeding programs.
Among the 75 common bean accessions evaluated, important sources of resistance to C. lindemuthianum, from Andean and Mesoamerican regions, were identified and have potential in breeding programs.
Furthermore, the identification of accessions resistant to C. lindemuthianum by the markers present new perspectives for the use of molecular marker assisted selection, making it more agile and effective selection process in breeding programs.
The authors have not declared any conflict of interests.
REFERENCES
|
Afanador LK, Haley SD, Kelly JD (1993). Adoption of a 'mini-prep' DNA extraction protocol for RAPD marker analysis in common bean (Phaseolus vulgaris L.). Ann. Rep. Bean Improv. Coop. 36:10-11.
|
|
|
|
Agarwal M, Shrivastava N, Padh H (2008). Advances in molecular marker techniques and their applications in plant sciences. Plant Cell. Rep. 27:617-631.
Crossref
|
|
|
|
Alzate-Marin AL, Costa MR., Arruda KM, Barros EG, Moreira MA (2003). Characterization of the anthracnose resistance gene present in Ouro Negro (Honduras 35) common bean cultivar. Euphytica 133:165-169.
Crossref
|
|
|
|
Alzate-Marin AL, Sartorato A (2004). Analysis of the pathogenic variability of Colletotrichum lindemuthianum in Brazil. Ann. Rep. Bean Improv. Coop. 47:241-242.
|
|
|
|
Balardin RS, Kelly JD (1998). Interaction between Colletotrichum lindemuthianum races and gene pool diversity in Phaseolus vulgaris. HortScience 123:1038-1047.
|
|
|
|
Beebe S, Skroch PW, Tohme J, Duque MC, Pedraza F, Nienhuis J (2000). Structure of genetic diversity among common bean landraces of Middle American origin based on correspondence analysis of RAPD. Crop Sci. 40:264-273.
Crossref
|
|
|
|
Beebe, S, Renjifo J, Gaitan E, Duque MC, Tohme J (2001). Diversity and origin of Andean landraces of common bean. Crop Sci. 41:854-862.
Crossref
|
|
|
|
Beraldo ALA, Colombo CA, Chiorato AF, Ito MF, Carbonell SAM (2009). Aplicação de marcadores SCARs para seleção de linhagens resistentes à antracnose em feijoeiro. Bragantia 68:53-61.
Crossref
|
|
|
|
Cárdenas F, Adams MW, Andersen A (1964). The genetic system for reaction of field beans (Phaseolus vulgaris L.) to infection by three physiologic races of Colletotrichum lindemuthianum. Euphytica 13:178-186.
|
|
|
|
Chacon, SMI, Pickersgill B, Debouck DG (2005). Domestication patterns in common bean (Phaseolus vulgaris L.) and the origin of the Mesoamerican and Andean cultivated races. Theor. Appl. Gen. 110:432-444.
Crossref
|
|
|
|
Coimbra-Gonçalves GK, Gonçalves-Vidigal MC, Coelho RT, Valentini G, Vidigal Filho PS, Lacanallo GF, Sousa LL, Elias HT (2016). Characterization and mapping of anthracnose resistance genes in mesoamerican common bean cultivar Crioulo 159. Crop Sci. 56:2904-2915.
Crossref
|
|
|
|
Corrêa RX, Costa MR, Good-God PI, Ragagnin VA, Faleiro FG, Moreira MA, Barros EG (2000). Sequence characterized amplified regions linked to rust resistance genes in the common bean. Crop Sci. 40:804-807.
Crossref
|
|
|
|
Damasceno e Silva KJ, Souza EA, Ishikawa FH (2007). Characterization of Colletotrichum lindemuthianum isolates from the State of Minas Gerais, Brazil. J. Phytopathol. 155:241-247.
Crossref
|
|
|
|
Ferreira JJ, Campa A, Perez-Vega E, Giraldez R (2008). Reaction of a bean germplasm collection against five races of Colletotrichum lindemuthianum identified in Northern Spain and implications for breeding. Plant Dis. 92:705-708.
Crossref
|
|
|
|
Geffroy V (1997). Dissection genétique de la résistance à Colletotrichum lindemuthianum, agent de l'anthracnose, chez deux génotypes représentatifs des pools géniques de Phaseolus vulgaris. (Doctoral Dissertation, Institute National Agronomique, Paris Grignon, 1997). France.
|
|
|
|
Geffroy V, Delphine S, Oliveira JCF, Sévignac M, Cohen S, Gepts P, Dron M (1999). Identification of an ancestral resistance gene cluster involved in the coevolution process between Phaseolus vulgaris and its fungal pathogen Colletotrichum lindemuthianum. Mol. Plant Microbe Interact. 12:774-784.
Crossref
|
|
|
|
Geffroy V, Sévignac M, Billant P, Dron M, Langin T (2008). Resistance to Colletotrichum lindemuthianum in Phaseolus vulgaris: a case study for mapping two independent genes. Theor. Appl. Genet. 116:407-415.
Crossref
|
|
|
|
Gepts P, Debouck D (1991). Origin, domestication, and evolution of the common bean (Phaseolus vulgaris L.). In. A. van Schoonhoven and O. Voysest O (ed). Common beans: Research for crop improvement. C.A.B. Intl., Wallingford, UK and CIAT, Cali, Colombia. pp. 7-53.
|
|
|
|
Gonçalves-Vidigal MC, Cruz AS, Lacanallo GF, Vidigal Filho PS, Sousa LL, Pacheco CMNA, Mcclean P, Gepts P, Pastor-Corrales MA (2013) Co-segregation analysis and mapping of the anthracnose Co-10 and angular leaf spot Phg-ON disease resistance genes in the common bean cultivar Ouro Negro. Theor. Appl. Genet. 126:2245-55.
Crossref
|
|
|
|
Gonçalves-Vidigal MC, Kelly JD (2006). Inheritance of anthracnose resistance in the common bean cultivar Widusa. Euphytica 151:411-419.
Crossref
|
|
|
|
Gonçalves-Vidigal MC, Lacanallo GF, Vidigal Filho PS (2008b). A new Andean gene conferring resistance to anthracnose in common bean (Phaseolus vulgaris L.) cultivar Jalo Vermelho. Plant Breed. 127:592-596.
Crossref
|
|
|
|
Gonçalves-Vidigal MC, Thomazella C, Vidigal Filho P.S, Kvitschal MV, Elias HT (2008a). Characterization of Colletotrichum lindemuthianum isolates using differential cultivars of common bean in Santa Catarina State, Brazil. Braz. Arch. Biol. Technol. 51:883-888.
Crossref
|
|
|
|
Guzman P, Gilbertson RL, Nodari R, Johnson WC, Temple SR, Mandala D, Mkandawire ABC, Gepts P (1995). Characterization of variability in the fungus Phaeoisariopsis griseola suggests coevolution with common bean (Phaseolus vulgaris). Phytopathology 85:600-607.
Crossref
|
|
|
|
Hernández-Godinez F, Gonzáles-Chavira M, Rodriguez-Guerra R, Acosta-Gallegos J, Simpson J (1998). La variación patogênica de Colletotrichum limdemuthianum y su importância em los programas de mejoramiento genético del frijol. Rev. Mex. Fitopatol. 16:63.
|
|
|
|
Kelly JD, Vallejo V (2004). A comprehensive review of the major genes conditioning resistance to anthracnose in common bean. HortScience 39:1196-1207.
|
|
|
|
Mahuku GS, Jara C, Cajiao C, Beebe S (2002). Sources of resistance to Colletotrichum lindemuthianum in the secondary gene pool of Phaseolus vulgaris and in crosses of primary and secondary gene pools. Plant Dis. 86:1383-1387.
Crossref
|
|
|
|
Mahuku GS, Riascos JJ (2004). Virulence and molecular diversity within Colletotrichum lindemuthianum isolates from Andean and Mesoamerican bean varieties and regions. Eur. J. Plant Pathol. 110:253-263.
Crossref
|
|
|
|
Melotto M, Coelho M.F, Pedrosa-Harand A, Kelly JD, Camargo, LEA (2004). The anthracnose resistance locus Co-4 of common bean is located on chromosome 3 and contains putative disease resistance-related genes. Theor. Appl. Genet. 109:690-699.
Crossref
|
|
|
|
Melotto M, Kelly JD (2000). An allelic series at the Co-1 locus for anthracnose in common bean of Andean origin. Euphytica 116:143-149.
Crossref
|
|
|
|
Melotto M, Kelly JD (2001). Fine mapping of the Co-4 locus of common bean reveals a resistance gene candidate, COK-4, that encodes for a protein kinase. Theor. Appl. Genet. 103:508-517.
Crossref
|
|
|
|
Mendéz-Vigo B, Rodríguez-Suárez C, Pa-eda A, Ferreira JJ, Giraldez R (2005). Molecular markers and allelic relationships of anthracnose resistance gene cluster B4 in common bean. Euphytica 141:237-245.
Crossref
|
|
|
|
Miklas PN, Kelly JD (2002). The use of MAS to develop pinto bean germplasm possessing Co-42 gene for anthracnose resistance. Ann. Rep. Bean Improv. Coop. 45:68-69.
|
|
|
|
Oblessuc PR, Francisco C, Melotto M (2015). The Co-4 locus on chromosome Pv08 contains a unique cluster of 18 COK-4 genes and is regulated by immune response in common bean. Theor. Appl. Genet. 128:1193-1208.
Crossref
|
|
|
|
Rodríguez-Guerra R, Ramírez-Rueda MT, Simpson J (2003). Variation in genotype, pathotype and anastomosis groups of Colletotrichum lindemuthianum isolates from Mexico. Plant Pathol. 52:228-235.
Crossref
|
|
|
|
Rodríguez-Guerra R, Ramírez-Rueda MT, Simpson J (2004). Capacidad de anastomosis de cepas del hongo Colletotrichum lindemuthianum (Sacc. et Magn.) Scrib., agente causal de la antracnosis del frijol (Phaseolus vulgaris L.). Rev. Mex. Fitopatol. 22:37-43.
|
|
|
|
Sansigolo AL, Gonçalves-Vidigal MC, Vidigal Filho PS, Gonela A, Kvitschal MV, Souza LL (2008). New races of Colletotrichum lindemuthianum in common bean (Phaseolus vulgaris L.) in Paraná State, Brazil. Ann. Rep. Bean Improv. Coop. 5:192-192.
|
|
|
|
Shiu SH. Bleecker AB (2001). Plant receptor-like kinase gene family: diversity, function and signaling. Sci. Signal. 113(re22):1-3.
Crossref
|
|
|
|
Sousa LL, Cruz AS, Vidigal Filho PS, Vallejo VA, Kelly JD, Gonçalves-Vidigal MC (2014). Genetic mapping of the resistance allele Co-52 to Colletotrichum lindemuthianum in the common bean MSU 7-1 line. Aust. J. Crop. Sci. 8:317-323.
|
|
|
|
Sousa LL., Gonçalves AO, Gonçalves-Vidigal MC, Lacanallo GF, Fernandez AC, Awale H, Kelly JD (2015). Genetic characterization and mapping of anthracnose resistance of common bean landrace cultivar Corinthiano. Crop Sci. 55:1025-1036.
Crossref
|
|
|
|
Trabanco N, Campa A, Ferreira JJ (2015). Identification of a new chromosomal region involved in the genetic control of resistance to anthracnose in common bean. Plant Genome 8:1-11.
Crossref
|
|
|
|
Vieira RF, de Paula Júnior TJ (2004). Importância do uso de sementes de feijão livres de patógenos. Informe Agropecuário 25:33-41.
|
|
|
|
Young RA, Melloto M, Nodari RO, Kelly JD (1998). Marker-assisted dissection of the oligogenic anthracnose resistance in the common bean cultivar, G 2333. Theor. Appl. Genet. 96:87-94.
Crossref
|


