Full Length Research Paper
ABSTRACT
Sorghum [Sorghum bicolor (L.) Moench], is believed to be originated in Ethiopia and Sudan. Although, many morphological and molecular diversity studies reveal the existence of genetic variations with sorghum populations, their distribution within basic races were not considered. Hence, the present study aimed to analyze the extent and distribution of genetic variation within basic Ethiopian sorghum landraces using SSR markers. A total of 107 landraces obtained from Ethiopian Biodiversity Institute (EBI) representing 12 ecological zones grouped according to their race types based on inflorescence and spiklet on field at their maturity time. Twelve SSR markers revealed a total of 110 alleles with average polymorphic content of 0.76 and the allele frequencies show 42 of them were rare (less than 0.05), 22 ranged from 0.05 to 0.1, while 46 of them were higher than 0.1. Expected and observed heterozygosity were 0.78 and 0.2, respectively. The genetic differentiation between populations were also moderate (FST=0.07 for races and 0.13 for E/zones) indicating continuous exchange of genes among them. Partitioning the total genetic variation also indicated 61.38 and 55.17% of the variations were among individuals within racial and zonal populations, respectively. Neighbor-Joining cluster analysis also indicated four major grouping of the landraces according to their racial groups where majority of race caudatum and durra form separate groups while intermediate durra-bicolor form two separate sub-clusters. Overall locus, the intra-racial population diversity showed the greatest genetic diversity (He=0.77 and 0.75) among race dura-bicolor and caudatum, respectively. Information with sorghum races along their important agronomic traits could be used for conservation and future breeding programs of sorghum.
Key words: Sorghum bicolor, races, genetic diversity, SSR.
Abbreviation: dNTPs, Deoxynucleotide triphosphates; EBI, Ethiopian Biodiversity Institute; NPGS, National Plant Germplasm System; PIC, polymorphic information content; PVP, polyvinylpyrrolidone; RAPD, random amplified polymorphic DNA; RFLP, restriction fragment length polymorphism; SSR, simple sequence repeats.INTRODUCTION
Sorghum [Sorghum bicolor (L.) Moench], a cultivated diploid (2n = 20) tropical cereal C4 grass plant, is the fifth most important cereal crop grown in the world. It is a monocotyledon plant of tropical origin, belonging to Poaceae family. Having nutritional composition similar to maize, starch is the major component of sorghum followed by protein, fat, and fiber (Council, 1996). The crop displays relatively high-water use efficiency compared to other cereals such as maize and wheat. Its wide adaptation to harsh environments, tolerance to stress conditions, diverse germplasm collections and its small genome size (710 Mb) made sorghum as an important botanical model crop for many tropical grasseswith complex genomes, which employ C4 photosynthesis. Sorghum is also the first crop genome of African origin to be sequenced (Dogget, 1965; Council, 1996).
Ethiopia is the second largest sorghum producer in Africa, after the Sudan and first among countries that have contributed many germplasm collections to the world collections of sorghum at both International Crop Research Institute for Semi-Arid Tropics (ICRISAT) and Griffin by National Plant Germplasm System (NPGS) (Demeke, 2013). It is one of the most important staple cereal crops after tef [Eragrostis tef (Zucc.) Trotter.] and maize (Zea mays L.) and holds third largest share of total cereal production with tef, maize, sorghum and wheat (Triticum aestivum L.) accounting for about 24.0, 16.8, 14.6, and 13%, respectively. Being an indigenous crop to Ethiopia, it is cultivated in almost all regions by subsistence farmers for various uses including as food and feed, house and fence construction, and prepare local beverages (CSA, 2014).
Though it was difficult to determine when and where sorghum domestication occurred, different studies suggested Ethiopia as a center of origin of sorghum due to the wide variation of the crop (McGuire, 2008; Vavilov, 1951). These also enable Ethiopian sorghum landraces as a source for an important agronomic trait including resistance to pest, sorghum midge (Contarinia sorghicola), and high lysine and protein contents (Fetene et al., 2011; Council, 1996). Sorghum had five basic races; namely bicolor, caudatum, durra, guinea and kafir. The entire races were differentiated morphologically based on their inflorescence, grain and glumes (Harlan and Wet, 1972). Clarissa et al. (2013) also described the geographic pattern of distribution of each race appears following the topography and climate variation present in Ethiopia. All the basic sorghum races except kafir also reported in Ethiopia. Accordingly, sorghum race durra is the main crop of the eastern highland region and mid elevation terrace of the north, while caudatum race is grown primarily in hot, dry valleys and lowland savannas in the south and west of Ethiopia. The intermediate race durra-bicolor predominates in the southwestern highland region, where cooler temperature and rain are higher than eastern and northern region. In contrast, bicolor and guinea races represent a very small part of Ethiopian sorghum diversity and both are mostly found in the Rift Valley region (House, 1985).
The diversity studies involving Ethiopian germplasms indicated the presence of huge genetic and morphological variations within their regions of origin and adaptation zones. Gebrekidan (1981) classified sorghum adaptation zones as: lowland (<1600 m above sea level (masl)), intermediate (1600-1900 masl) and highland (>1900 masl) in Ethiopia (Gebrekidan, 1981). Qualitative and quantitative studies, in addition to RAPD, AFLP and SSR markers utilized by different scholars also revealed the same amounts of variations among their collections (Ayana and Bekele, 1998; Geleta et al., 2006; Cuevas and Prom, 2013). SSR marker studies from Eritrean, Eastern Kenya, Benin and Zambia collections also showed presence of wide genetic diversity of sorghum bicolor in Africa mainly in Eastern regions (Tesfamichael et al., 2014; Catherine et al., 2016; Antoine et al., 2015; Ng'uni et al., 2011). The existence of imbalanced sorghum races in the sample collections of the different studies from Ethiopia might contribute to the overall observed genetic variation (Cuevas and Prom, 2013). Ethiopian Biodiversity Institute (EBI) collected large numbers of farmers’ landraces though they do not have a racial category. Antoine et al. (2015) also recommended research on genetic diversity to integrate both botanical races and morpho-physiological characteristics of the crops for better preservation of sorghum genetic resources. In addition, morphological studies involving germplasms from Ethiopia and Eritrea show the greatest share of variation observed were carried by their panicle compactness and shape, which is 31% (Ayana and Bekele, 1998). Therefore, racially partitioned diversity studies among founding major basic races and representative of the whole collections of Ethiopian adaptation zones were lacking. Hence, the present study aimed at analyzing the genetic variation within basic Ethiopian sorghum races.
MATERIALS AND METHODS
Germplasm collections
The accessions used for this study were landrace accessions collected by EBI. A total of 107 sorghum landrace accessions were selected based on phonological evaluation of inflorescence and spiklet types at their maturity time in order to define the racial classifications in 2015/2016 cropping season at Arsi Negelle Research Station based on their passport data. The materials were received and planted along with other germplasm by Melkassa Sorghum Improvement Program. In addition to the difference in head morphology, geographical distribution of the sorghum races across the country were considered for selection of their adaptation zones. All basic sorghum races except kafir and from the intermediate types, widely distributed durra-bicolor were included. The selected 107 sorghum landrace collections were grouped based on their source of origins into 12 populations, which each contained 9 landrace accessions for DNA extraction and genotyping study.
DNA extraction and PCR amplification
The seeds of collected sorghum genotypes were planted at National Agricultural Biotechnology Research Center (NABRC), Holetta, on seedling tray in greenhouse for germination. Genomic DNA was extracted from 2-week old seedlings using fresh leaves according to Xin et al. (2003) utilizing only two ordinary buffers that made the genomic DNA available for PCR amplification reactions. Approximately 50 mm2 single leaf sample per landraces was harvested to PCR plates for their DNA extraction. The two buffers include: Buffer A made from 100 mM NaOH and 2% Tween 20, which are made fresh from their stock solutions (10M NaOH and 20% Tween 20) and Buffer B consisting of 100 mM Tris-HCl and 2 mM EDTA, whose pH set to 2.0. Once the buffers are ready, the genomic DNA was extracted with the following procedures: (1) Approximately 30 mm2 leaf tissue transferred to 96-well plates; (2) 50 µL buffer A added and incubated for 10 min at 95°C in thermo cycler; (3) 50 µL buffer B was added and mixed at moderate speed; (4) Aliquot PCR mixture to 96-well plates at a reaction volume of 20 µL/well; (5) and finally transfer approximately 1.5 µL DNA from the crude DNA plates to PCR plates with a 96-pin applicator (Figure 1).

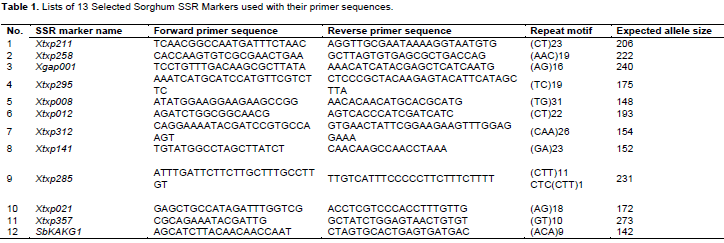
Twelve polymorphic SSR primer pairs (Table 1) were selected for genotyping the selected 107 sorghum landraces. PCR amplification was carried out in 20 µL reaction volume containing 1.5 µL crude genomic DNA, 2.25 µl PCR buffer with MgCl2 (17.5 mM), 1.8 µl of dNTPs (10 mM), 0.45 µl each of forward and reverse primers (10 mM), 0.133 µl of Taq Polymerase (5U), 0.1% BSA (Bovine Serum Albumin) (w/v) and 1% PVP (w/v). The amplifications were carried out with thermo cycler programmed for initial denaturation at 94°C for 15 min, the second denaturation at 94°C for 30 s, annealing at 50°C for 1 min, extension at 72°C for 2-min, final elongation at 72°C for 20 min and holding temperature at 4°C until conclusion. PCR products were analyzed by loading the 3 µl PCR products along with a 3 µL loading dye mixed with Gel Red (at a ratio of 1000:1) using 3.5% agarose gel electrophoresis run with 100 V for 3 h along with DNA Ladder (500 bp bioline Hyperladder V).
Data collection and statistical analysis
Once gel images were taken with Gel documentation, the PCR fragments were scored manually by estimating their base pair size as compared with known fragments size ladders that were run gel electrophoresis along with each accession. The number of alleles (N), major allele frequency (A), observed heterozygosity (Ho), expected heterozygosity/gene diversity (He) and polymorphism information content (PIC) for each SSR locus were analyzed using PowerMarker 3.25 (Liu and Muse, 2005).
Pairwise genetic distance was calculated as given by Nei and Takezaki (1994). Further, the allelic data were subjected to estimate the genetic distances using simple matching coefficients and the genotypes were clustered using Neighbor Joining method. Both the clustering analysis and PCoA were done using DARwin software ver. 6.0.13. The data were tested for presence of population structure and analysis of molecular variance (AMOVA) was performed to separate the total molecular variance into components between groups, within groups and intra population variation using Arlequin version 3.0 software (Excoffier et al., 2005). Pairwise genetic differentiation between different groups was assessed with fixation index (Weir and Clark, 1984) as implemented in Arlequin software.
RESULTS AND DISCUSSION
Marker polymorphism
A total of 110 alleles were separated using 12 SSR markers (Table 2). The number of alleles per marker ranged from 5 (xtxp298 and xtxp258) to 18 (xtxp211) with an average of 9.2 alleles per locus. Xtxp298 and xtxp012 hold the lowest and highest number of genotypes (6 and 19, respectively) with an average of 15.8 genotypes per locus. The allele frequencies varied from 0.005 for marker xtxp285 to 0.648 for xtxp312 with an overall average frequency of 0.109. The mean number of alleles per locus observed in this study was higher than similar SSR studies with accessions from North Eastern Benin, 7 (Antoine et al., 2015), Zambia, 4.4 (Ng'uni et al., 2011), Eastern Kenya, 5.05 (Catherine et al., 2016), Eritrea, 4.8 (Tesfamichael et al., 2014) and Egypt, 7.3 (El-Awady et al., 2008), and Ethiopian collections in combination with other countries (Agrama and Tuinstra, 2003). However, it is lower than Cuevas and Prom (2013) population structure and diversity study for 137 Ethiopian germplasm conserved at USDA-ARS National Plant Germplasm System, that is 14 per locus. Out of the total 110 alleles, specifically 42 alleles had frequencies below 0.05 (rare alleles), 22 alleles had a frequency within 0.05 to 0.10 (common alleles) while the rest 46 alleles had frequency higher than 0.10 becoming an abundant allele. While across all races average number of frequencies ranged from 7.17 in intermediate durra-bicolor to 4.75 in race bicolor whereas mean gene diversity ranged from 0.32 (bicolor) to 0.22 (durra). Their mean number of alleles within the different races ranged from 7.17 (durra-bicolor) to 4.75 (bicolor). Likewise, their gene diversity ranged from 0.77 for durra-bicolor to 0.70 for durra, guinea and bicolor (not shown).
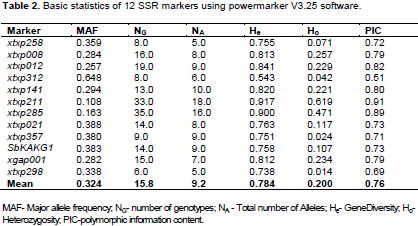
Their polymorphic information content (PIC) varied from 0.51 (xtxp312) to 0.91 (xtxp211) with an average of 0.76 and the expected and observed heterozygosity (gene diversity and heterozygosity respectively) ranged from 0.54 (xtxp312) to 0.92 (xtxp211) and 0.014 (xtxp298) to 0.62 (xtxp211), respectively.
Higher polymorphism within the present Ethiopian landraces observed may be indication of the extensive and regular seed exchange farming system within farmers of Ethiopia (Mcguire, 2000). This form of seed migrations also adds allelic variations to landraces avoiding genetic drift. Thus, the observed rare alleles could be useful as an additional source of important agronomic traits. In fact, Sorghum, a genus having evolved across a wide range of environments in Africa, exhibits a great range of phenotypic diversity and numerous resistances to abiotic and biotic stresses (Dogget, 1965). It is cultivated in all regions of Ethiopia from 400 to 2500 masl. Hence, the wider agro-ecological diversity of Ethiopian climates from where the samples were collected and the presence of wider morphological variations observed within them might contribute to its genetic variations (Mcguire, 2000).
Similar findings by Cuevas and Prom (2013), and to a certain extent Agrama and Tuinstra (2003) also reported average PIC values 0.78 and 0.622, respectively. However, the observed PIC value is higher than that of Geleta et al. (2006), Antoine et al. (2015) and Catherine et al. (2016) who reported 0.46, 0.33 and 0.49, respectively. This may be the result of low numbers of accessions considered in their studies and sample collections represented are from specific areas of agro-ecologies. Although sorghum is considered as self-pollinating species, cross-pollinations between sorghum landraces are believed to be as high as 7%, and can even reach 70% in certain races in particular environments. The observed high allelic frequencies could also arise from outcrossing within wild and weedy relatives (House, 1985; Dogget, 1965).
AMOVA analysis
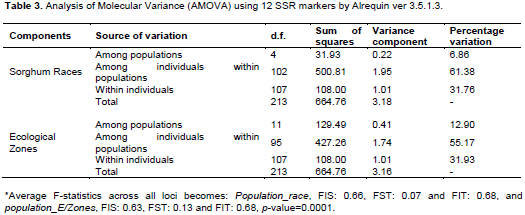
Partitioning the total variation of 107 Ethiopian sorghum landraces using 12 SSR markers revealed the presence of 61.38 and 55.17% variations explained by individual differences within race and their ecological zones, respectively. In contrast, the variations among the two populations are very small (6.86% among races and 12.9% among zones). A considerable amount of its total variation was recorded across the overall individual landraces that is 31.7% with a moderate degree of gene differentiation among racial populations in terms of allele frequencies, FST: 0.073 (Table 3). The moderate genetic differentiation among the present populations in terms of allele frequency also indicated the continuous exchange of genes between them. This finding also supports earlier studies by Cuevas and Prom (2013) who found genetic differentiation of 0.10 among 137 Ethiopian sorghum maintained at NPGS. However, Ganapathy et al. (2012) reported high estimate of fixation index (FST=0.35, P=0.001) using 82 Indian genotypes.
Cluster analysis and pairwise genetic dissimilarity
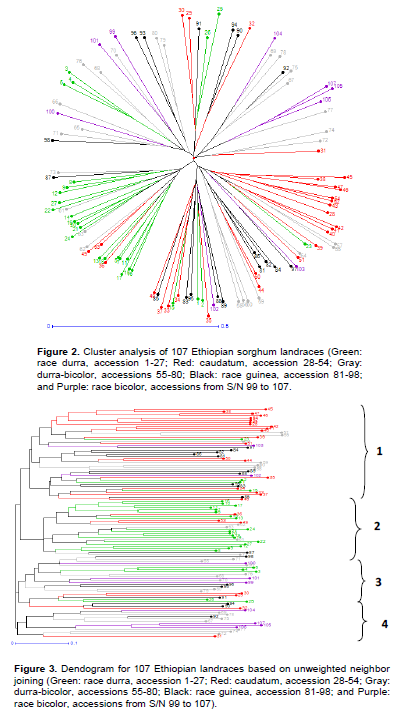
Neighbor-joining analysis indicated four major clusters (Figures 2 and 3). The first cluster, representing the largest numbers of accessions of all races in scattered manner, formed two sub-clusters inside; one with mainly of caudatum and another uniformly intermixed race. The second cluster most uniquely contained mainly durra race (18/27, 67% of the total population representing the race) along with rare numbers of caudatum and durra-bicolor (3 and 4). Exceptionally, no bicolor race clustered under this group, while only a single guinea represented. Like the first cluster, the third cluster also formed two sub-clusters and most intermediate durra-bicolor structured in one sub-cluster along with other race types. Under the final cluster 4, majority of race durra-bicolor contained along with a single caudatum and guinea race, and rare number (that is 4) of bicolor. RFLP analysis on 94 accessions also reported the greatest amount of diversity within races bicolor and guinea when racially classified. They also reported the race bicolor appeared highly variable and did not form a specific group. Hence, it was believed to be distributed wherever sorghum is grown (Wang et al., 2013). The most unique and clustered race in PCoA, race durra, is abundant in Ethiopian and Sudan as well and Harlan and Wet (1972) also reported settlers in warm highlands of Ethiopia have used the durra sorghum as their foundation of their agricultural system almost 500 years ago. These may be the reason why durra-bicolor intermediate race were also abundant in the country. In addition, caudatum, a race being adapted to harsh conditions, are found most commonly in areas receiving 250 to 1,300 mm of rain annually (Stemler et al., 1977). The bicolor and guinea, representing the smallest parts of Ethiopian diversity, distributed evenly across all clusters except the bicolor race which did not form a group within durra race cluster.
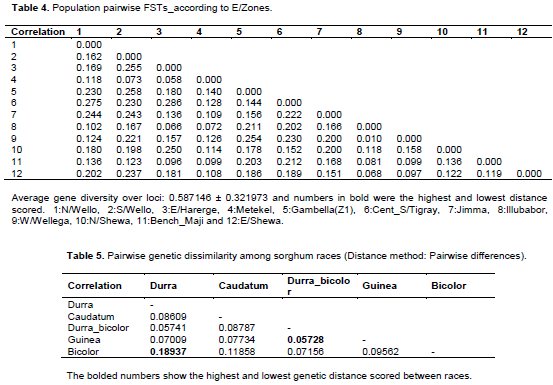
Matrix of pairwise genetic distance of the racial population relationships (Table 5) indicated the existence of the highest dissimilarity between race bicolor and durra (highest genetic distance, 0.19) while the lowest score was registered between intermediate durra-bicolor and guinea (0.06). Whereas, among populations of different ecological zones pairwise genetic distance ranged from 0.0096 to 0.286 between West Wollega and Illubabor, and between Central and South Tigray and East Harerge, respectively. The least genetic distance between West Wollega and Illubabor (0.0096) may be due to the close proximity of the two zones where free seed exchange might occur. In contrast, the greatest dissimilarity (0.286) recorded were between central and south Tigray and east Harerghe zones (Table 4).
CONCLUSION AND RECOMMENDATIONS
In general, the racial classification among S. bicolor could be used for in-situ and ex-situ conservation and genetic dissimilarity with their respective agronomic characteristics favors the future crops germplasm breeding programs. In this regard, Ethiopian sorghum races were structured into four major clusters according to their racial difference except for race bicolor and guinea. Bicolor found being scattered within other groups. The greatest genetic distance found between bicolor and caudatum while between the Ethiopian zones, central and south Tigray and east Harerghe. There was also a huge variation observed among the populations of both racial classification and ecological zones (61.38 and 55.17%, respectively). The greater mean number of alleles per locus (9.2) and PIC value of 0.76 in the present study also indicate the presence of high genetic diversity among Ethiopian sorghum collections and the discriminatory power of the selected markers. There is also a moderate levels of genetic differentiation among races (FST= 0.07) and Ethiopian sorghum producing zones (FST=0.13). Hence, racial groups could also be used as representation of the germplasm collection along with the commonly known diverse agro-ecological and zonal collections and their adaptation zones.
In line with the present study, the future research areas should include molecular studies along with the morphological components using markers linked to specific agronomic traits to enable the use of racial groupings within sorghums in its breeding areas. In addition, classification of the national sorghum germplasm collections according to their race was also needed since the Ethiopian landraces have been used as the source of important traits.
CONFLICT OF INTERESTS
The authors have not declared any conflict of interests.
ACKNOWLEDGEMENTS
The authors thank the technical staff at National Agricultural Biotechnology Research Center, Holetta for generously helping technically throughout the time in their laboratory and Melkassa management staff’s in easing the work with financial issues.
REFERENCES
|
Agrama HA, Tuinstra MR (2003). Phylogenetic diversity and relationships among sorghum accessions using SSRs and RAPDs. African Journal of Biotechnology 2(10):334-340. |
|
|
Antoine AM, Hubert AS, Paulin S, Clement A, Corneille A, Rollande AD (2015). Genetic diversity of Sorghum bicolor (L.) Moench) landraces from Northwestern Benin as revealed by microsatellite markers. African Journal of Biotechnology 14(16):1342-1353. |
|
|
Ayana A, Bekele E (1998). Geographical patterns of morphological variation in sorghum (Sorghum bicolor (L.) Moench) germplasm from Ethiopia and Eritrea: Qualitative characters. Hereditas 129(3):195-205. |
|
|
Catherine WM, Reuben MM, Duncan TK, Steven MR, Arthur K (2016). Genetic variability of sorghum landraces from lower Eastern Kenya based on simple sequence repeats (SSRs) markers. African Journal of Biotechnology 15(8):264-271. |
|
|
Clarissa TK, Jef AD, Stephen K (2013). The Genepool of Sorghum bicolor and its improvements. In A. H. Paterson (Ed.), Genomics of the Saccharinae, Plant Genetics and Genomics:Crops and Models (pp. 23-39). Texas, USA: Springer. |
|
|
Council NR (1996). Lost crops of Africa. Volume 1, Grains. Washigton, DC: National Academy Press. |
|
|
CSA (2014). Report on area and production of major crops: private peasant holdings of Meher season (Vol. 1). Addis Ababa, Ethiopia. |
|
|
Cuevas HE, Prom LK (2013). Assessment of molecular diversity and population structure of the Ethiopian sorghum (Sorghum bicolor (L.) Moench) germplasm collection maintained by the USDA-ARS National Plant Germplasm using SSR markers. Genetic resources and Crop Evolution 60(6):1817-1830. |
|
|
Demeke MDF (2013). Analysis of Incentives and Disincentives for Sorghum in Ethiopia. Rome: MAFAP, FAO. |
|
|
Dogget H (1965). Disruptive section in crop development. Nature 4981:279-280. |
|
|
El-Awady M, Youssef SS, Selim EEM, Ghonaim MM (2008). Genetic diversity among Sorghum bicolor genotypes using simple sequence repeats (SSRs) markers. Arab Journal of Biotechnology 11(2):181-192. |
|
|
Excoffier L, Laval G, Schneider S (2005). Arlequin (version3.0): An integrated software package for population genetics data analysis. Switzerland: 3012 Berne. |
|
|
Fetene M, Okori P, Gudu S, Mneney EE, Tesfaye K (2011). Delivering New Sorghum and Finger Millet Innovations for Food Security and Improving Livelihoods in Eastern Africa. Kenya, Nairobi: ILRI. |
|
|
Ganapathy KN, Gomashe SS, Rakshit S, Ambekar SS, Ghorade RB, Biradar BD, Patil JV (2012). Genetic diversity revealed utility of SSR markers in classifying parental lines and elite genotypes of sorghum (Sorghum bicolor L. Moench). Australian Journal of Crop Science 6(11):1486-1493. |
|
|
Gebrekidan B (1981). Salient features of the sorghum breeding strategies used in Ethiopia. Ethiopian Journal of Agricultural Sciences 3:97-104. |
|
|
Geleta N, Labuschagne MT, Viljoen CD (2006). Genetic diversity analsis in sorghum germplasm as estimated by AFLP, SSR and morpho-agronomical markers. Biodiversity and Conservation 15(10):3251-3265. |
|
|
Harlan JR, Wet JM (1972). Simplified classification of cultivated sorghum. Crop Science 12(2):172-176. |
|
|
House LR (1985). A Guide to Sorghum Breeding (Second ed.). India: International Crops Research Institute for the Semi-Arid Tropics. |
|
|
Liu K, Muse S (2005). PowerMarker: integrated analysis environment for genetic marker data. Bioinformatics 21:2128-2129. |
|
|
Mcguire S (2000). Farmers' management of sorghum diversity in Eastern Ethiopia. In C Almekinders, W deBoef (Eds.), Encouraging Diversity: The Conservation and Development of Plant Genetic Resources (pp. 43-48). London: Intermediate Technology. |
|
|
McGuire SJ (2008). Securing access to seed: Social relations and Sorghum seed exchange in Eastern Ethiopia. Human Ecology 36(2):217-229. |
|
|
Nei M, Takezaki N (1994). Estimation of Genetic Distances and Phylogenetic Trees from DNA Analysis. In Proceedings of the 5th World Congress on Genetics applied to Livestock production, Vol.21 (pp. 405-412). Ontario, Canada: University of Guelph. |
|
|
Ng'uni D, Geleta M, Bryngelsson T (2011). Genetic diversity in sorghum (Sorghum bicolor (L.) Moench) accessions of Zambia as revealed by simple sequence repeats (SSR). Hereditas 148(2):52-62. |
|
|
Stemler A, Harlan J, de Wet J (1977). The sorghums of Ethiopia. Economic Botany 31:446-460. |
|
|
Tesfamichael T, Githiri SM, Kasili RW, Skilton R, Solomon M, Nyende A (2014). Genetic diversity analysis of Eritrean sorghum (Sorghum bicolor (L.) Moench) germplasm using SSR markers. Molecular Plant Breeding 5(13):1-12. |
|
|
Vavilov N (1951). The Origin, Variation, Immunity and Breeding of Cultivated Plants. USA: The Chronica Botanica Co. |
|
|
Wang YH, Upadhyaya HD, Burrell AM, Sahraeian SM, Klein RR, Klein PE (2013). Genetic structure and linkage disequilibrium in a diverse representative collection of the C4 model plant, Sorghum bicolor. Genetics 3:783-793. |
|
|
Weir BS, Clark C (1984). Estimating F-Statistics for the analysis of population structure. Evolution 38(6):1358-1370. |
|
|
Xin Z, Velten J, Oliver M, Burke J (2003). High-throughput DNA extraction method suitable for PCR. Biotechniques 34(4):820-824. |
|
Copyright © 2024 Author(s) retain the copyright of this article.
This article is published under the terms of the Creative Commons Attribution License 4.0