ABSTRACT
Pseudomonas putida is a well-known plant growth promoting bacterium (PGPB) with the capacity to improve plant growth. The objective of this study was to assess the effect of P. putida towards plant growth and its interaction with its microbial ecosystem. P. putida strain PF1P was isolated from the rhizosphere of banana plant (Musa species). The isolated P. putida strain PF1P possess ability to secrete acetic acid (42.1 µg/mL) and gibberellic acid (10.1 µg/mL). Further study conducted showed that P. putida strain PF1P was able to increase 54.6% of fresh plant weight and 51.3% of root length for Brassica chinensis var parachinensis. Soil microbial assessment indicates that P. putida strain PF1P did not have any detrimental effects as Shannon diversity index showed an increase in the microbial diversity from 2.635 to 2.903 with the colony forming unit per gram of soil increase from 9 × 107 to 5 × 109, respectively for control and treated soil. Soil health check also showed that inoculation of P. putida strain PF1P does not cause any negative effect to the soil. Inoculation of P. putida strain PF1P proved to improve the growth of the plant and microbial diversity in the soil. Mass production of this bacterium should be studied for possible industrial application.
Key words: Encapsulation, auxin, gibberellic acid, plant growth promoting bacteria.
Bacteria are present abundantly in soil and interact with the environment surrounding them for their survival. The availability of these bacteria is crucial for healthy plant growth where they provide beneficial conditions to the plant (Pandya and Desai, 2014). Some of the important conditions where they exert their beneficial roles are through production of phytohormones (such as auxin and gibberellic acid), siderophores and antimicrobial metabolites (Iqbal and Hasnain, 2013). Pseudomonas, Bacillus, Enterobacter and Arthrobacter species are among the genera of common plant growth promoting bacteria (PGPB) that are often studied and explored (Mohite, 2013; Pandya and Desai, 2014; Reetha et al., 2014). Among these, Pseudomonas spp. is more popular studied for its ability to produce auxin and gibberellic acid.
Both plant hormones are crucial for normal development, especially in root growth. Auxin also known as indole-3-acetic acid (IAA), is an important plant hormone in the regulation of plant development and production of longer roots and increased the number of root hairs which will directly increase nutrient uptake by the plant (Mohite, 2013). On the other hand, gibberellic acid (GA) is widely documented to be involved in seed germination, seedling emergence, flower and fruit growth and also promotion of root growth and root hair abundances (King and Evans, 2003; Sponsel, 2001).
From the diversity of Pseudomonads studied, Pseudomonas putida is well-known for its agriculture usage (Xie et al., 1996; Pattern and Glick, 2002; Rodriguez et al., 2014). For example in Euphorbia pulcherrima cultivars and Brassica napus, P. putida was found to have positively affected plant growth (Cheng et al., 2012; Rodriguez et al., 2014). In addition to the study on the usage, researchers had also studied the feasibility of encapsulating these bacteria to enhance its functionality and its survival rate (Rekha et al., 2007).
With the huge potential of using P. putida as in agriculture, this study proposes to further its functionality on Brassica chinensis var parachinensis (Choy sum) a common vegetable dish for most Chinese in Malaysia. In addition, the benefits to the microbial ecosystems will be explored.
Samples collection and isolation of Pseudomonas spp.
Six soil samples were collected randomly from the MARDI Organic Farm. The soil was taken at 15 cm from the rhizosphere of the selected plants. The soil was kept in a zip lock bag and process upon reaching laboratory. Ten grams of the soil was mixed with 100 mL of sterile distilled water (sH2O). The soil suspension was agitated vigorously at 250 rpm for 2 h. Hundred and fifty microliter of the soil suspension was pipetted and dispensed onto King’s B Agar. The agar plates were incubated at 30 ± 2°C for 48 h. Emerging colonies from the agar plates were picked and sub-cultured onto fresh media plates.
Screening of IAA and GA3 producing Pseudomonas spp.
For indole-3-acetic acid estimation, method used by Iqbal and Hasnain (2013) was followed with modification. Bacterial cell suspension was incubated at 30 ± 2°C for 48 h and the culture broth was adjusted to 106 CFU/mL prior to pipetting of 2 mL culture broth into Eppendorf tube and centrifuged at 10,000 rpm for 30 min. After centrifugation, 1 mL of the supernatant and 2 mL of Salkowski’s reagent were mixed properly and placed in dark for 30 min. Development of pink color indicated IAA production and the amount of IAA was measured using Nanodrop spectrophotometer at wavelength of 530 nm. A standard curve with different concentrations was constructed using synthetic IAA (Sigma) used to determine the amount of IAA produced by each Pseudomonas spp.
Screening of GA3 producing Pseudomonas spp. was done according to the method proposed by Panday and Desai (2014). Standard curve was constructed using synthetic GA3 (Sigma) and measured using Nanodrop spectrophotometer at 254 nm.
Identification and characterization potential of Pseudomonas spp.
Isolation of DNA was conducted using Qiagen DNeasy DNA extraction protocol as suggested by the manufacturer. Polymerase chain reaction (PCR) parameter and conditions used for this study followed the optimization made by Jeffrey (2008) with the modification on the primers used. The 16S rRNA gene region was amplified using universal primers F8 (AGA GTT TGA TCM TGG CTC) and rP2 (ACG GCT ACC TTG TTA CGA CTT). PCR products obtained were subjected to purification using Vivantis GF-1 Gel DNA recovery kit following the protocol provided by the manufacturer. The purified PCR products were sent for sequencing at First Base Laboratories Sdn. Bhd., Selangor. Results obtained from the sequencing were then compared with the databases from National Center for Biotechnology Information (NCBI). Biochemical characterization of Pseudomonas spp. was conducted using BIOLOG Gen III microplate according to the method described by the manufacturer (BIOLOG Inc.).
Preparation of alginate beads
The selected bacteria were grown in a 2 L conical flask containing 1 L of nutrient broth for 24 h with vigorous agitation at 250 rpm. Upon 24 h, the CFU/mL of bacteria were adjusted to 108 CFU/mL before the suspension was centrifuged at 10,000 rpm for 30 min. Cell pallet obtained was then washed with sH2O to remove the excess nutrient broth before addition of the cell into 1 L of 3% (w/v) sodium alginate solution. The suspension was then stirred for 30 min to ensure proper mixing. The suspension was then dripped into 0.1 M calcium chloride (CaCl2) solution. The beads formed were left in the CaCl2 solution for 1 h before being washed with sH2O to remove excess salt form during the process. The beads were then kept in 4°C until used.
Glasshouse trial
Two seeds were planted in each pot with 5 replicates and 2 cycles. Treatments were Pseudomonas spp. strain PF1P and control. Five pots were supplied with 2 g of Pseudomonas spp. strain PF1P beads every 15 days until harvested. B. chinensis var parachinens was harvested at 35 days after planting. The fresh and dry weight and root length of the plants for each treatment were recorded. The test was conducted in a randomized completely block design (RCBD).
Soil microbial activity and soil health evaluation
Each similar treatment was pooled together and mixed evenly; 100 g of the soil were taken and proceed with BIOLOG Eco Plate analysis according to the manufacturer’s protocol (Biolog Inc.). Shannon’s diversity index (H) and substrate richness (S) were calculated to determine the effect of the microbe application towards the microbial community in the soil. Shannon’s diversity index was calculated based on the equation of H= Σ(Pi x lnPi), where by Pi is the proportional optical density of each wells at 96 h and S was calculated based on the number of different carbon substrate utilized by the microbial community (Wang et al., 2007). From the pooled soil, 500 g was scooped into ziploc bags and sent for analysis of its cation exchange capacity (CEC), conductivity, total organic matter and percentage of organic carbon.
Statistical analysis
Data were statistically analyzed with t-test at 95% confidence level.
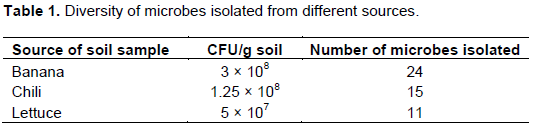
Isolation of Pseudomonas spp.
A total of 50 Pseudomonas spp. were isolated from the 6 soil samples collected from 3 different rhizospheres of MARDI organic farm (Table 1). It was observed that banana rhizosphere contained the most population of bacteria compared to lettuce and chili (Table 1). Soil sample from lettuce recorded the lowest CFU/g and total number of bacteria isolated.
Screening for phytohormones activities, identification and characterization of Pseudomonas spp.
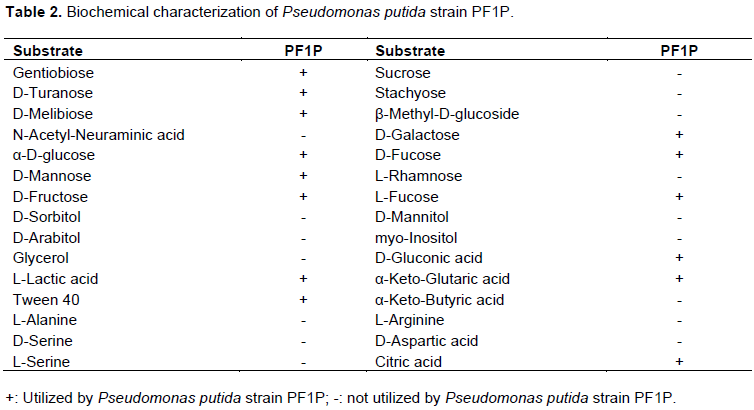
From the 50 isolates of bacteria tested, 30% of the bacteria produced IAA and 35% GA3. It was observed that Pseudomonas spp. strain PF1P gave the highest IAA and GA3 reading compared to the other Pseudomonas spp. isolated. Pseudomonas spp. Strain PF1P was later identified as P. putida based on their 16S rRNA gene sequence. P. putida strain PF1P was observed to utilize several carbon sources such as gentiobiose, D-turanose, D-melibiose, α-D-glucose, D-mannone, D-fructose, L-fucose, L-lactic acid, Tween 40, D-galactose, D-fructose, L-fucose, D-gluconic acid, and α-keto-glutaric acid and citric acid (Table 2).
Efficacy of P. putida strain PF1P on glasshouse trial
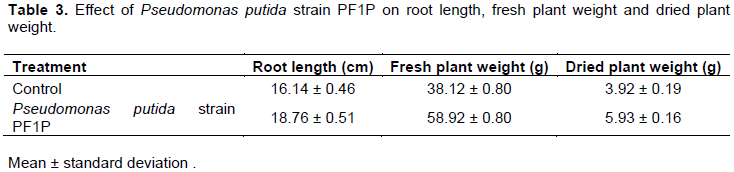
From the trial conducted, it was observed that P. putida strain PF1P increased the fresh weight of the B. chinensis var parachinensis by 54.6% and encouraged the elongation of roots by 51.3%. T-test conducted showed significant results on the increased of B. chinensis var parachinensis weight and root elongation at the confidence level of 95%. The average fresh weight, dry weight and root length are shown in Table 3.
Effect of P. putida strain PF1P on soil microbial activity and soil health
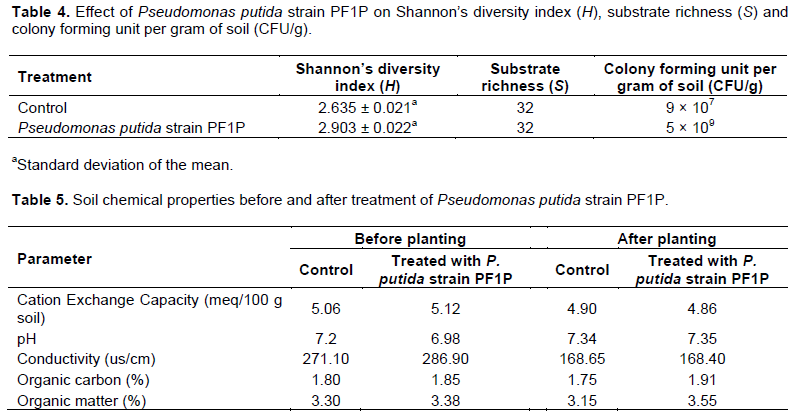
It was observed that P. putida strain PF1P did not have any detrimental effect on the soil health when the results are compared to the control. Shannon’s diversity index (H) calculated for soil inoculated with P. putida strain PF1P was observed to be 2.903 as compared to the control of only 2.635 (Table 4). Soil treated with P. putida strain PF1P showed higher CFU (5 × 109 CFU/g) compared to control of only 9 × 107 CFU/g (Table 4). There are no significant differences in the substrate richness (S) for both soil inoculated with P. putida strain PF1P and control. It was observed that organic matter and organic carbon content for treated soil sample increased by 5 and 3% respectively as compared to non-treated soil. The pH reading for treated soil did not show any significant changes similar to results obtained for CEC, conductivity and pH, respectively (Table 5).
IAA and GA3 producer by Pseudomonas
In this study, P. putida strain PF1P showed a slightly higher IAA secretion of approximately 42.1 µg/mL as compared to studies done by Pattern and Glick (2002) where they only recorded IAA secretion of 32.7 µg/mL. In another study conducted by Reetha et al. (2014), Pseudomonas fluorescens isolated from the rhizosphere of onion produced only 15.38 µg/mL of IAA. According to Pattern and Glick (2002), the high reading of IAA content was correlated to the accumulation of indolepyruvic acid which reacted with the Salkowski’s reagent. Puga-Freista et al. (2012) however observed that production of auxin was closely related to the host plants and growth of other soil organisms such as earthworms.
P. putida strain PF1P which was isolated from banana rhizosphere gave a GA3 reading of 10.1 µg/mL. This result was slightly higher than the results obtained by Verma et al. (2016) from P. putida (9.8 µg/mL) and P. flourescens (6.64 µg/mL) isolated from Cuddalore district in India. Ambawade and Pathade (2015) noted that production of high gibberellic acid from Bacillus siamensis BE 76 was related to the host plant. In the study, B. siamensis BE 76 isolated from banana rhizosphere gave better GA3 production. This might be due to secretion of certain stimulus by the banana plants that was not present in other plant.
Efficacy of P. putida strain PF1P on glasshouse trial
The increase of B. chinensis var parachinensis fresh weight as shown in Table 3 might be due to the increase in the absorption of nutrients and water by the root system of the plant. This could be caused by the colonization of the bacteria on the roots and the rhizosphere that encourage the absorption of minerals by plants through the development of more elongated and nested root structure (Rodriguez et al., 2014). This result correlates with P. putida strain PF1P, which caused a 51.3% increase in root elongation. Furthermore, treated B. chinensis var parachinensis showed the formation of plenty of root hairs at the primary and secondary roots. Study done by Pattern and Glick (2002) also supported the role of P. putida in increasing the rooting system of Vigna radiate.
Effect of P. putida strain PF1P on the soil microbial activity
Shannon’s diversity index showed that soil treated with P. putida strain PF1P gave higher microbial activities and diversity as compared to the control. These phenomena could be classified as “rhizosphere effect” whereby colonization of P. putida strain PF1P could have secreted certain compounds in the plant rhizosphere and enhanced different microbial populations in the ecosystem (Rekha et al., 2007). This was observed in the increase of CFU/g count in soil inoculated with P. putida strain PF1P (Table 4).
P. putida strain PF1P isolated from the rhizosphere of banana plot was observed to be producing both IAA and GA3. Encapsulated P. putida strain PF1P was able to enhance the rooting system and soil microbial diversity in glass house test. P. putida strain PF1P was also found to be non-detrimental to its microbial ecosystem and could be formulated with other fertilizers to reduce the dependency on chemical fertilizers.
The authors have not declared any conflict of interests.
REFERENCES
|
Ambawade MS, Pathade GR (2015). Production of gibberellic acid by Bacillus siamensis BE 76 isolated from banana plant (Musa spp). Int. J. Sci. Res. 4(7):394-398.
|
|
|
|
Cheng Z, Woody OZ, McConkey BJ, Glick BR (2012). Combined effects of the plant growth-promoting bacterium Pseudomonas putida UW4 and salinity stress on the Brassica napus proteome. Appl. Soil Ecol. 61:255-263.
Crossref
|
|
|
|
Iqbal A, Hasnain S (2013). Auxin producing Pseudomonas strains: Biological candidates to modulate the growth of Triticum aestivum beneficially. Am. J. Plant Sci. 4:1693-1700.
Crossref
|
|
|
|
Jeffrey LSH (2008). Isolation, characterization and identification of actinomycetes from agriculture soils at Semongok, Sarawak. Afr. J. Biotechnol. 7(20):3700-3705.
|
|
|
|
King RW, Evans LT (2003). Gibberellins and flowering of grasses and cereals: prising open the lid of the "Florigen" black box. Annu. Rev. Plant Physiol. Plant Mol. Biol. 54:307-328.
Crossref
|
|
|
|
Mohite B (2013). Isolation and characterization of indole acetic acid (IAA) producing bacteria from rhizospheric soil and its effect on plant growth. J. Soil Sci. Plant Nutr. 13(3):638-649.
Crossref
|
|
|
|
Pandya ND, Desai PV (2014). Screening and characterization of GA3 producing Pseudomonas monteilii and its impact on plant growth promotion. Int. J. Curr. Microbiol. Appl. Sci. 3(5):110-115.
|
|
|
|
Pattern CL, Glick BR (2002). Role of Pseudomanas putida indo lactic acid in development of the host plant root system. Appl. Eniron. Microbial. 68:3795-3801.
Crossref
|
|
|
|
Puga-Freitas R, Abbad S, Gigon A, Garnier-Zarli E, Blouin M (2012). Control of Cultivable IAA-Producing Bacteria by the Plant Arabidopsis thaliana and the Earthworm Aporrectodea caliginosa. Applied and Environmental Soil Science 2012.
Crossref
|
|
|
|
Reetha S, Bhuvaneswari G, Thamizhiniyan P, Ravi Mycin T (2014). Isolation of indole acetic acid (IAA) producing rhizobacteria of Pseudomonas fluorescens and Bacillus subtilis and enhance growth of onion (Allim cepa.L). Int. J. Curr. Microbiol. Appl. Sci. 3(2):568-574.
|
|
|
|
Rekha PD, Lai WA, Arun AB, Young CC (2007). Effect of free and encapsulated Pseudomonas putida CC-FR2-4 and Bacillus subtilis CC-pg104 on plant growth under gnotobiotic conditions. Bioresour. Technol. 98:447-451.
Crossref
|
|
|
|
Rodriguez RZ, Matson MVC, Montiel LGH, Amador BM, Puente ER, Lara L (2014). Effect of Pseudomonas putida on growth and anthocyanin pigment in two Poinsettia (Euphorbia pulcherrima) cultivars. The Scientific World Journal, 2014.
|
|
|
|
Sponsel VM (2001). The deoxy xylulose phosphate pathway for the biosynthesis of plastidic isoprenoids: early days in our understanding of the early stages of gibberellin biosynthesis. J. Plant Growth Regul. 20:332-345.
Crossref
|
|
|
|
Verma P, Yadav AN, Khannam KS, Mishra S, Kumar S, Saxena AK, Suman A (2016). Appraisal of diversity and functional attributes of thermotolerant wheat associated bacteria from the peninsular zone of India. Saudi Journal of Biological Sciences.
Crossref
|
|
|
|
Wang GH, Jin J, Chen XL, Liu JD, Liu XB, Herbert SJ (2007). Biomass and catabolic diversity of microbial communities with long-term restoration, bare fallow and cropping history in Chinese Mollisols. Plant Soil Environ. 53(4):177-185.
|
|
|
|
Xie H, Pasternak JJ, Glick BR (1996). Isolation and characterisation of mutants of the plant growth- promoting rhizobacterium Pseudomonas putida GR- 122 that overproduce indole acetic acid. Curr. Microbiol. 32:67-71.
Crossref
|