ABSTRACT
This study aimed to determine the genetic diversity of antibiotics and plants extract resistant Staphylococcus aureus using molecular technique. A total of 106 human clinical samples were collected from patients in three different hospitals within Ekiti State. Microbiological and molecular analyses were performed using standard methods. Antimicrobial susceptibility test with common antibiotics and plant leaves extracts was carried out using disk and agar well diffusion methods. Urine samples from male patients recorded high percentage of S. aureus (57.1%) as compared to the female patients (43.1%). Percentage of S. aureus recorded from the ear swab samples in male patients (58.3%) was also high as compared to the female (41.7%). From the nose swab samples, female patients recorded 60% as compared to their male counterpart (40%). Equal percentage (50%) of wound infection caused by S. aureus was observed in both male and female patients. Of the eight antibiotics used in this study, the S. aureus isolated were susceptible to ofloxacin (25 to 36 mm), gentamycin (16 to 23 mm) and erythromycin (11 to 25 mm). All the isolates confirmed resistance to ceftaxidime and cloxacillin. Nine isolates were susceptible to cefuroxime with zones of inhibition that ranged from 10 to 25 mm, while 11 were susceptible to ceftriaxone with zone of inhibition between 11 and 20 mm. Only one isolate was sensitive to augmentin (amoxicillin/clavulanate) with zone of inhibition of 20 mm. Out of three plant extracts used in this study, aqueous leaf extract of Terminalia catappa demonstrated highest antibacterial activity on the test isolates with zone of inhibition of 16 to 36 mm followed by Mangifera indica with inhibition ranging from 11 to 32 mm, while least inhibition ranging from 11 to 20 mm was exhibited by Acalypha wikesiena. Random amplified polymorphic DNA (RAPD) proved to be useful as genetic markers in determining genetic diversity among antibiotic and plant extract resistant S. aureus.
Key words: Acalypha wikesiena, multidrug-resistant strains, Staphylococcus aureus, genetic diversity.
Modern medicine is no doubt faced with an unprecedented threat posed by multidrug-resistant bacteria (Levy and Marshall, 2004). It has been advised that a multidisciplinary approach is required to tackle the emergence and evolution of drug resistance microorganisms (Martinez and Baquero, 2002). In their study, Baquero et al. (2003) averred that the multidisciplinary approach will be a challenge given that the trend of antibiotic resistance appears as a reflection of the characteristic of a dynamic, highly complex, and self-organizing system evolving at an instance of complex situations. New molecular technologies promise to offer adequate and effective tools in the fight against antibiotic resistance strains of microorganisms. Advance molecular approaches have also been reported to be effective containment of multidrug-resistant strains, given the accurate detection of bacterial species from different environment. Currently, the main advantage of molecular resistance testing is the rapid turn-around time and improved accuracy, both of which are relevant particularly in cases of life-threatening diseases such as meningitis and sepsis, for which rapid detection, identification and resistance testing is important (Johansen et al., 2003). It is noteworthy to add that, rapid polymerase chain reaction (PCR)-based assays for resistance testing have been used as excellent complementary tools in the laboratory for the detection of methicillin-resistant Staphylococcus aureus (MRSA) (Warren et al., 2004).
Medicinal plants as alternative therapeutic agent have recently attracted the attention of the biological scientific communities. Use of medicinal plants involves the isolation and identification of secondary metabolites produced by plants and is used as active principles in medicinal preparations. Application of various herbal medicinal preparations for the treatment of S. aureus associated illnesses in Nigeria has been reported (Akinyemi et al., 2005). Volumes of reports have demonstrated the antimicrobial properties of several of these plants against multi-drug resistant bacteria, particularly methicillin resistant S. aureus (Ankri and Mirelman, 1999; Adesino et al., 2011; Khan et al., 2016).
Methicillin resistant S. aureus (MRSA) is a bacterium responsible for several difficult-to-treat infections in humans. MRSA is any strain of S. aureus that has developed through the horizontal gene transfer and natural selection, multi-resistance to beta-lactam antibiotics, which include the penicillins (methicillin, dicloxacillin, nafcillin, oxacillin, etc.) and the cephalosporins. Understanding the S. aureus lineages therefore, is crucial in controlling nosocomial infections. Recent studies on S. aureus in Nigeria have revealed an escalating burden of MRSA. However, the S. aureus genotypes circulating among hospitalized patients in hospitals found in Ekiti State are not known, hence this study.
Sampling
In this research, a total of 106 human clinical samples were collected and transported within 2 h of collection to the Microbiology Research Laboratory in the Department of Biological Sciences, Afe Babalola University, Ado-Ekiti for microbiological analysis. The samples used in this research were urine, wound, nose and ear swabs obtained from patients hospitalized at different wards of Ekiti State University Teaching Hospital (EKSUTH), State Specialist Hospital Ikere-Ekiti, and General Hospital Iyin-Ekiti in Ekiti State Nigeria. Samples were obtained from both male and female of different ages. From men, 33 urine samples, 14 ear swabs, four nose swabs and seven wound swabs were obtained. On the other hand, a total of 45 samples were obtained from women comprising 25 urine samples, ten ear swabs, six nose swabs and seven wound swabs. Each urine sample was aseptically collected into a sterile sampling bottle, while ear, nose and wound swabs were collected using a sterile swab stick.
Plant sample collection and preparation
Fresh plant leaves of Terminalia catappa (Almond), Mangifera indica (Mango) and Acalypha wikesiena (Acalypha) were obtained within the premises of Afe Babalola University and identified in the Department of Botany, University of Ibadan, Nigeria. Voucher specimens for T. catappa (UIH 22567), M. indica (UIH 22568) and A. wikesiena (UIH 22569) were deposited at University of Ibadan Herbarium of the Department of Botany. The leaves were brought to the laboratory, stripped from their stems, washed with distilled water and crushed into a fine infusion using a laboratory mortar and pestle. The solution was filtered with No. 1 Whatman filter paper and finally passed through membrane filter to obtain sterile extract. These were labelled and kept at 4°C in the refrigerator for use. A clean filter was used to obtain the extracts respectively into three well labelled beakers.
Qualitative analysis of phytochemical constituent
Simple standard chemical tests as previously described by Harborne (1973) and Chethana et al. (2013) were employed for detecting the presence of some phytochemical components such as saponins, tannins, alkaloids, steroids, phenol, terpenoid, flavonoid and glycosides in the plant extract.
Isolation and identification of test organisms
The isolates gotten from the various sites were identified using standard microbiological method as described by Cheesbrough (2000).
Antibiotic sensitivity testing
Antimicrobial susceptibility of the isolates was determined by the Kirby Bauer disc diffusion method according to CLSI guidelines (2012) using the following antibiotics: ofloxacin (OFL), augmentin (AUG), ceftaxidime (CAZ), cefuroxime (CRX), gentamycin (GENT), ceftriaxone (CTR), erythromycin (ERY) and cloxacillin (CXC). Young culture of the isolate was suspended in a sterile saline solution (0.85% NaCl). The suspension was adjusted to match 0.5 McFarland turbidity standards. The antibiotics discs were aseptically placed on Mueller Hinton agar surface using sterile forceps and incubate at 37°C for 24 to 48 h. Effectiveness of the antibiotics was shown by clear zones around them which were measured and interpreted as either resistance, intermediate, or susceptible.
Plant extract inhibitory test
Agar well diffusion method was used as previously described (Magaldi et al., 2004; Valgas et al., 2007) to evaluate the antimicrobial activity of the plants extracts. Clear zones of inhibition after incubation were measured as previously described (Lino and Deogracious, 2006).
DNA isolation and purification of bacterial genomic DNA
Sterile dry swabs were used for streaking clinical samples onto sterile nutrient agar plates (CONDA). Inoculated plates were incubated at 37°C for 48 h. Stock cultures of the pure isolates were prepared and refrigerated at 4°C to be kept for further analysis. Extraction and purification of genomic DNA of the bacterial isolates was carried out using the ZR Fungal/Bacterial DNA MiniPrep™ (Zymo). The procedure employed was as described by the manufacturer.
RAPD PCR analysis
RAPD PCR analysis used in this study was performed as previously described by Onasanya et al. (2003). DNA primers tested were purchased from Operon Technologies (Alemada, Califonia, USA) with each being 10 nucleotides long. Two separate concentrations of DNA, 24 and 96 ng per reaction were used to test reproducibility as well as eliminate irregular amplification products from the analysis. Thirteen primers were screened for their ability to amplify the DNA of the S. aureus isolate. Six out of the 13 screened primers were found useful because they showed polymorphism. These were used for amplification of DNA of the 20 isolates. Amplification were performed in 25-µl reaction mixture consisting of genomic DNA, IX reaction buffer, (Promega), 100 µM each of dATP, dCTP, dGTP and dTTP, 0.2 µM Operon random primer, 2.5 µM MgCl2 and 0.2 μl (1 unit) of Taq polymerase (Boehringer, Germany). A single primer was used in each reaction. Amplification was performed in a thermowell microtiter plate (Costa Corporation) using a Perkin Elmer programmable Thermal Controller model 9600. The cycling program was set as follows: initial denaturation at 94°C for 3 min followed by 45 cycles each at 94°C for 1 min, 40°C for 1 min., 72°C for 2 min and final extension at 72°C for 7 min. Amplified DNA products were analyzed by electrophoresis in 1.4% agarose gel using TAE buffer (45 Mm Tris-acetate, 1 Mm EDTA, pH 8.0) at 100 V. Gels were visualized by staining with ethidium bromide solution (0.5 µg/ml) and banding patterns were observed using the Photogel documentation system.
Phylogenetic analysis
Positions of unequivocally scorable RAPD bands were transformed into a binary character matrix (to indicate the presence or absence of a band at a particular position). Pair-wise distance matrices were compiled by the Numerical Taxonomy System (NTSYS) 2.0 software (Rohfl, 2000) using a simple matching coefficient for estimation by means of the Jaccard’s coefficient to construct a similarity matrix (Ivchenko and Honov, 1998). Cluster analysis and dendrogram were produced on the basis of the unweighted pair group method arithmetic (UPGMA) (Sneath and Sokal, 1973; Jako et al., 2009) as the banding profiles of the six RAPD primers represent a novel approach for rapid screening of the multidrug resistant.
Statistical analysis
Data generated were expressed in mean ± standard error mean (SEM). One-way analysis of variance (ANOVA) followed by Student T-test (Walson, 1989; AOAC 1990) was used for this research.
All samples analyzed for the presence of Staphylococcus on mannitol salt agar and nutrient agar media showed obvious bacterial growths. Out of 106 collected samples from both male and female patients of different age brackets admitted to the three hospitals within Ekiti State, Ekiti State University Teaching Hospital (EKSUTH) had the highest number of patient colonized with S. aureus.
From urine samples, high percentage of S. aureus in male patients (57.1%) as compared to their female counterpart (43.1%) was recorded. Percentage of S. aureus recorded from the ear swab samples in male patients (58.3%) was also high as compared to the female (41.7%). From the nose swab samples, female patients recorded 60% as compared to their male counterpart (40%). Equal percentage (50%) of wound infection caused by S. aureus was observed in both male and female patients.
Further to this study, 20 representatives of the S. aureus isolates were picked for multidrug resistance pattern to antibiotics and plant leaves aqueous extracts of which 9 were obtained from patients’ urine, three from the noses, four from the ears and four from wounds. These isolates were code-numbered accordingly. All the isolates were completely resistant to CAZ and CXC as there was no zone of inhibition observed. Nine isolates were observed to be sensitive to CRX with zones of inhibition ranging from 10 to 25 mm, while 11 isolates were sensitive to CTR with range between 11 and 20 mm. Meanwhile, only one isolate was sensitive to AUG 20 mm zone of inhibition (Figure 1). However, the 20 S. aureus isolates used in the study were all susceptible to OFL with clear zones of inhibition ranging from 24 to 35 mm followed by GEN 16 to 23 mm and ERY 11 to 25 mm.
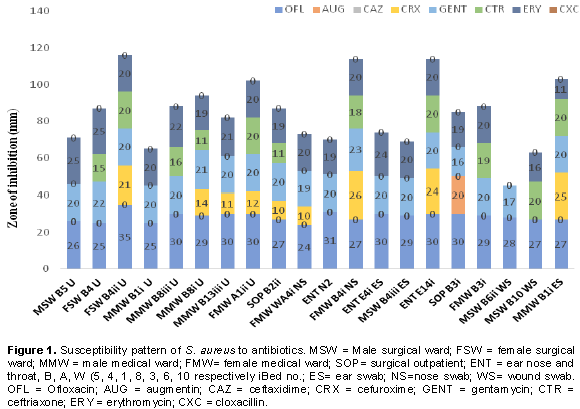
This apparently high level of susceptibility of the isolates to OFL suggest that ofloxacin (OFL) could be a drug of choice for treating infections caused by S. aureus in the study area, especially at the present time, when S. aureus strains are resistant to other commonly used antibiotics as reported in many literatures. This finding is consistent with previous reports of Amadi et al. (2007) and Chigbu and Ezeronye (2003). Also, 100% sensitivity of S. aureus isolates to ofloxacin (OFL) was reported by Obi et al. (1996). In addition, high inhibition of OFL against S. aureus isolates was also highlighted by Chalita et al. (2004). However, low sensitivity was reported among most strains of methicillin resistant S. aureus (MRSA) isolated from patients with ocular infections (Uwaezuoke and Aririatu, 2004). S. aureus susceptibility to gentamycin (GEN) in this present study compares favourably with reports published in the South-western part of Nigeria, by Paul et al. (1982) and Ndip et al. (1997).
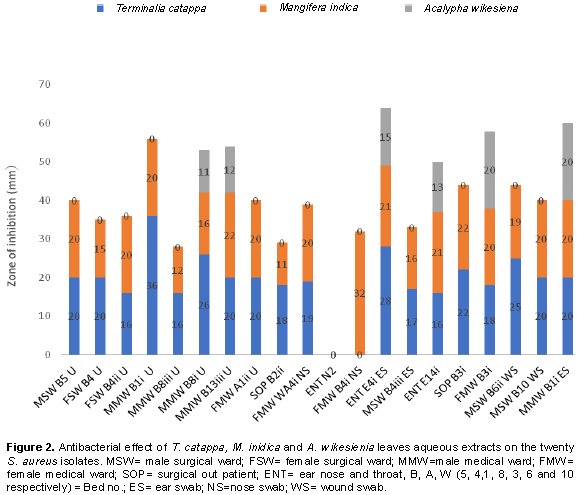
On the other hand, the sensitivity pattern of 20 S. aureus isolates to leaves extract shows that T. catappa demonstrated the highest level of effectiveness against 18 isolates which included isolates gotten from urine, ear, nose and wounds with zone of inhibition that ranges between 16 to 36 mm, with 2 isolates resistant to the plant. Although, 19 isolates were inhibited by M. indica, their zone of inhibition ranged between 11 and 32 mm, with only one isolate resistant and 14 out of the 20 isolates showed resistance to the aqueous leaf extract of A. wikesiena. Susceptibility of 6 isolates to A. wikesiena ranged from 11 to 20 mm zones of inhibition (Figure 2). The highest antimicrobial activity was observed with T. catappa which had better effect on 18 isolates with wider zones of inhibition than all the commercial antibiotics used in this study. Also, aqueous leaf extract of M. indica demonstrated higher antimicrobial effect on 19 isolates and its inhibitory activity was better than all the antibiotics used except OFL from which a wider zone of inhibition was observed. This indicates that the extracts of the leaf are as effective as the commercially prepared antibiotics thus confirming its potency in the treatment of S. aureus-mediated infections as well as gastroenteritis and skin dermatitis. Moreover, since the leaf extracts exhibited inhibitory activity on the test organisms, the antibacterial activity appears to be broad spectrum (Oluduro et al., 2011). The phytochemicals detected in the leaves aqueous extracts of T. catppa, M. indica and A. wikesiena in this study corresponds to earlier studies carried out with other plants by Onah et al. (2002), Akpan and Usoh (2004) and Oyewole et al. (2008).
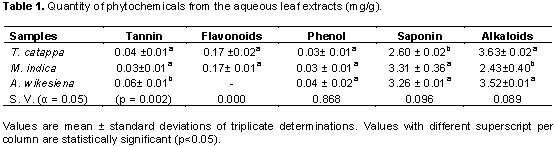
The quantity of tannin present in T. catappa was significantly different from the quantity of tannin present in A. wikesiena but not significantly different from the value obtained in M. indica (Table 1). Furthermore, flavonoids content in T. catappa shows no significant difference from that in M. indica. No significant difference existed in the phenol quantity between T. catappa, M. indica and A. wikesiena. Also, the quantity of saponin in T. catappa was not significantly different from the values of M. indica and A. wikesiena (Table 1).
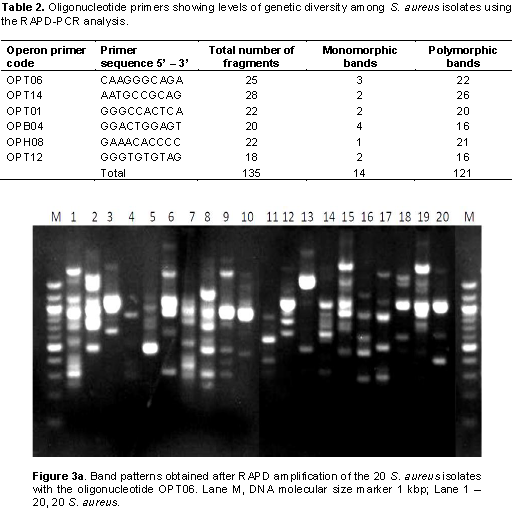
In this study, it was observed that all isolates from patients’ samples receiving treatment from EKSUTH expressed similar pattern in their resistance to the antibiotic and plant extract used. Genetic fingerprinting of the 20 S. aureus isolates from urine, ear, nose and wound swabs of patients receiving treatment from 3 hospitals within Ekiti State was compared using RAPD-PCR markers. Of the 13 arbitrary primers tested, six primers (Table 2) showed a good level of polymorphism and reproducibility. The amplification reaction of 6 oligonucleotide primers yielded 135 bands of which 121 were polymorphic with sizes ranging between 200 and 3,000 bp (Figure 3a to f). 121 polymorphic RAPD markers were used to construct the phylogenetic relationship dendrogram among 20 S. aureus isolates into different group’s genotypes and sub groups depending on their reaction with antibiotics and plants extract used in the study.
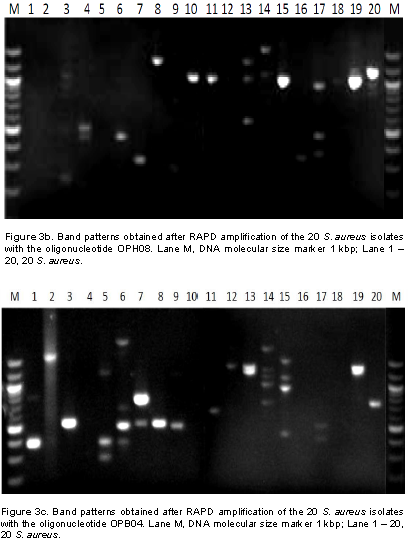
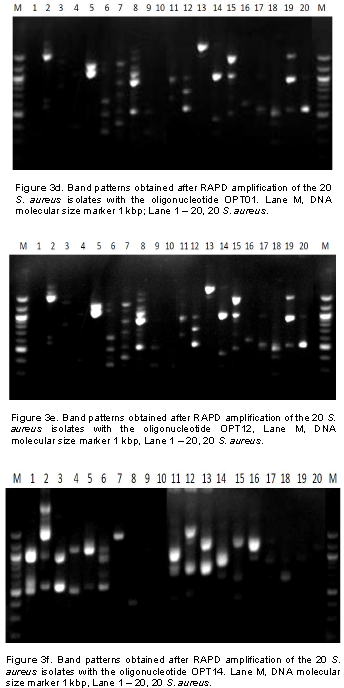
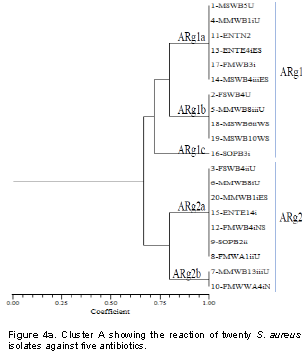
From the reaction of 8 antibiotics on the 20 S. aureus isolates, two different reactive groups emerged which consists of antibiotics reaction group 1 (ARg1) and antibiotics reaction group 2 (ARg2) at 75% similarity (Figure 4a). ARg1 yielded 3 subgroups which include ARg1a, ARg1b and ARg1c. ARg1a consisted of six isolates (1-MSWB5U, 4-MMWB1iU, 11-ENTN2, 13- ENTE4iES, 17-FMWB3i and 14-MSWB4iiiES). The reactions of these isolates with five antibiotics were identical. ARg1a consisted of four isolates (2-FSWB4U, 5-MMWB8iiiU, 18-MSWB6iiWS and 19-MSWB10WS) that were also identical in their reaction with 5 antibiotics. ARg1c had only 1 isolate (16-SOPB3i). ARg2 yielded only 2 subgroups consisting of ARg2a and ARg2b. ARg2a consisted of 7 isolates (3-FSWB4iiU, 6-MMWB8iU, 20-MMWB1iES, 15-ENTE14i, 12-FMWB4iNS, 9-SOPB2ii and 8-FMWA1iiU). These isolates show similar pattern of activities in their reaction with the 5 antibiotics. ARg1a consisted of two isolates (7-MMWB13iiiU and 10-FMWWA4iNS). Both isolates in ARg2b were identical in their reaction with the five antibiotics.
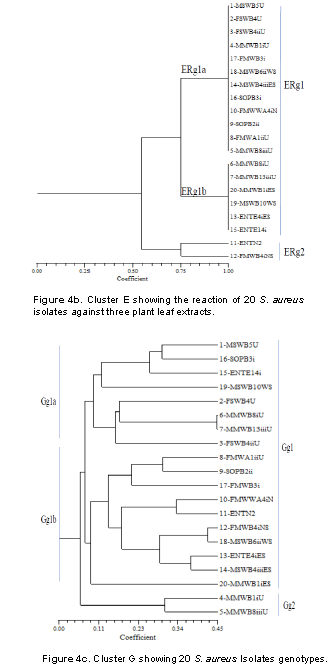
Figure 4b presents cluster E that shows the reaction of 3 plant leaf extracts (T. cattapa, M. indica and A. wikesiena) on 20 S. aureus isolates. At 55% coefficient of similarity, 2 major groups were formed, namely ERg1 and ERg2. ERg1 developed two subgroups namely ERg1a and ERg1b. ERg1a which consists of 12 isolates (1-MSWB5U, 2-FSWB4U, 3-FSWB4iiU, 4-MMWB1iU, 17-FMWB3i, 18-MSWB6iiWS, 14-MSWB4iiiES, 16-SOPB3i, 10-FMWWA4iNS, 9-SOPB2ii, 8-FMWA1iiU and 5-MMWB8iiiU). These isolates reacted in the same way to the aqueous leaf extracts. ERg1b consisted of six isolates (6-MMWB8iU, 7-MMWB13iiiU, 20-MMWB1iES, 19-MSWB10WS, 13-ENTE4iES and 15-ENTE14i) which also reacted in the same way with the extracts. ERg2 had two isolates (11-ENT N2 and 12-FMWB4iNS) which reacted differently to the extracts.
Figure 4c presents cluster G which consists of two major group genotypes (Gg) represented as Gg1 and Gg2 at 10.5% similarity. Gg1 yielded 2 subgroups namely Gg1a and Gg1b at 11% similarity. Gg1a further gave rise to two subgroups consisting of Gg1ai and Gg1aii; Gg1b gave rise also to two subgroups Gg1bi and Gg1bii as with 11.5% similarity. Gg2 did not yield any obvious subgroups. However, the (Gg) cluster formed three distinct groups; the first group consisted of four isolates (1, 16, 15 and 19), the second group consisted of four isolates (2, 6, 7 and 3) while the third group consisted of 9 isolates (8, 9, 17, 10, 11, 12, 18, 13 and 14). From the first group, it was observed that only the group genotype of isolate 1 was resistant to the antibiotics, making the group an antibiotic resistance emergent group. Group genotypes of the isolates in group 2 revealed antibiotics susceptibility because no resistance was found in the isolates.
Molecular research studies indicate that resistance is a complex system, resulting from multiple interacting mechanisms at the genomic, regulatory and expression levels. If the complementary effort of both the culture-dependent and molecular technique as evidenced in this study is anything to go by, molecular assays for resistance testing will not replace conventional culture-based antibiotic susceptibility testing in the immediate future (Fluit et al., 2001).
According to Adesida et al. (2007), identification of antibiotic resistant organism using molecular techniques such as 16s rRNA, random amplification of polymorphic DNA (RAPD) and plasmid profile causing infectious processes proves to be very essential for effective antimicrobial and supportive therapy.
By virtue of this analysis using molecular method (RAPD- PCR), it was possible to group isolates of similar genotypes with the same resistance behaviour to antibiotics and plants extract. Although major bands from RAPD reactions are highly reproducible, challenges like minor bands proves difficult to repeat due to random priming nature of PCR reaction and potential confounding effects associated with co-migration with other markers.
The authors have not declared any conflict of interests.
REFERENCES
|
Association of Official Analytical Chemists (AOAC) (1990). Official Methods of Analysis of the AOAC, 15th Edn. Washington, D.C. P 365.
|
|
|
|
Adesida SA, Abioye OA, Bamiro BS (2007). Associated risk factorsand pulsed field gel electrophoresis of nasal isolates of Staphyloccous aureus from medical students in a tertiary hospital in Lagos, Nigeria. Brazilian Journal of Infectious Diseases 11(1):63-69.
Crossref
|
|
|
|
|
Adesino GO, Jibo S, Aggu VE. Ehinmidu JO (2011). Antibacterial activity of fresh juices of Allium cepa and Zingiber ofiicinale against multidrug resistant bacteria. International Journal of Pharmacy and Biological Sciences 2:289-295.
|
|
|
|
|
Akinyemi KO, Oladapo O, Okwara C E, Ibe CC, Fasure KA (2005). Screening of crude extracts of six medicinal plants used in South-West Nigerian unorthodox medicine for anti-methicillin resistant Staphylococcus aureus activity. BMC Alternative Med. 5:6-13.
Crossref
|
|
|
|
|
Akpan EJ, Usoh IF (2004). Phytochemical screening and effect of aqueous root extract of Raphiahookeri (raffia palm) on metabolic clearance rate of ethanol in rabbits. Biochemistry 16(l):37-42.
|
|
|
|
|
Amadi ES, Nwofor GE, Ogbu O, Ayogu TE, Ononiwu CE (2007). Resistance of Staphylococcus aureus to commonly used antibiotic obtained from Different sources in Abakaliki. African Journal of Science and Technology 8(1):1728-1739.
|
|
|
|
|
Ankri S, Mirelman D (1999). Antimicrobial properties of allicin from garlic. Microbes and Infection 1:125 -129.
Crossref
|
|
|
|
|
Ayoola GA, Coker HB, Adesegun SA, Adepoju-Bello AA, Obaweya K, Ezennia EC, Atangbayila TO (2008). Phytochemical screening and antioxidant activities of some selected medicinal plants used for malaria therapy in southwestern Nigeria. Tropical Journal of Pharmaceutical Research 7:1019-1024.
|
|
|
|
|
Baquero F, Coque TM, Canton R (2003). Antibiotics complexity and evolution. ASM News 69:547-552.
|
|
|
|
|
Chalita MK, Hofling-Lima AL, Paranhos A, Schor P, Belfort R (2004). Shifting trends in vitro antibiotic susceptibilities for common ocular isolates during a period of 15 years. American Journal of Ophthalmology 137(1):43-51.
Crossref
|
|
|
|
|
Cheesbrough M (2003). Medical Laboratory Manual. Tropical Health Technology, Low Priced Edition. Doddington, Cambridge shire, England. pp. 132-143.
|
|
|
|
|
Chethana GS, Hari Venkatesh KR, Giopinath SM (2013). Preliminary phytochemicals analysis of Clerodendrum inerme. International Research Journal of Pharmacy 4(5):208-209.
Crossref
|
|
|
|
|
Chigbu, CO, Ezeronye OU (2003). Antibiotic resistant Staphyloccous aureus in Abia State of Nigeria. African Journal of Biotechnology 2(10):374-378.
Crossref
|
|
|
|
|
Clinical and Laboratory Standards Institute (CLSI) (2012). Performance Standards for Antimicrobial Disk Susceptibility Tests, Approved Standard, 7th ed., CLSI document M02-A11. 950 West Valley Road, Suite 2500, Wayne, Pennsylvania 19087, USA.
|
|
|
|
|
Fluit AC, Visser MR, Schmitz FJ (2001). Molecular detection of antimicrobial resistance. Clinical Microbiology Reviews 14:836-871.
Crossref
|
|
|
|
|
Harborne JB (1973). Phytochemical Methods. Chapman and Hall Ltd, London. pp. 149-188.
|
|
|
|
|
Jako E, Ari E, Ittzes P, Horvath A, Podani J (2009). BOOL-AN: A method for comparative sequence analysis and phylogenetic reconstruction. Molecular Phylogenetics and Evolution 52:887-897.
Crossref
|
|
|
|
|
Johansen IS, Lundgren B, Sosnovskaja A, Thomsen VO (2003). Direct detection of multidrug-resistant Mycobacterium tuberculosis in clinical specimens in low- and high-incidence countries by line probe assay. Journal of Clinical Microbiology 41:4454-4456.
Crossref
|
|
|
|
|
Khan R, Baeshen MN, Saini KS, Bora RS, Al-Hejin AM, Baeshen NA (2016). Antibacterial activities of Rhazyastricta leaf extracts against multidrug resistant human pathogens. Biotechnology & Biotechnological Equipment 30:1016-1025.
Crossref
|
|
|
|
|
Levy SB, Marshall B (2004). Antibacterial resistance worldwide: causes, challenges and responses. Nature medicine 10:S122-S129.
Crossref
|
|
|
|
|
Lino A, Deogracios O (2006). The in-vitro antibacterial activity of Annona senegalensis, Securidacca longipendiculata and Steanotaenia araliacea-Ugandan Medicinal plants. African Health Sciences 6(1):31-35.
|
|
|
|
|
Magaldi, S., Mata-Essayag, S. and Hartung, C. de Capriles (2004), Well diffusion for antifungal susceptibility testing. International Journal of Infectious Diseases 8 pp. 39-45.
Crossref
|
|
|
|
|
Martinez JL, Baquero F (2002). Interactions among strategies associated with bacterial infection: pathogenicity, epidemicity, and antibiotic resistance. Clinical Microbiology Reviews15:647-679.
Crossref
|
|
|
|
|
Mboso OE, Eyong EU, Odey MO, Osakwe E (2013). Comparative phytochemical screening of Ereromastax speciosa and Ereromastax polysperma. Journal of Natural Product and Plant Resources 3(2):37-41.
|
|
|
|
|
Ndip, RN, Ebah LME, Onile BA (1997). Antibiogram of Staphylococcus aureus from clinical syndromes in Ilorin, Nigeria. Journal of Medical Laboratory Science 6:24-26.
|
|
|
|
|
Obi CL, Iyiegbuniwe AE, Olukoya DK, Babalola C, Igunbor EO, Okonta A. (1996). Antibiogram and plasmids of Staphylococcus aureus and coagulase negative staphylococci isolated from different clinical sources. Central African Journal of Medicine 42(9):258-261.
|
|
|
|
|
Olajide R, Akinsoyinu AO, Babayemi OJ, Omojola AB, Abu AO, Afolabi KD (2011). Effect of processing on energy values, nutrient and anti-nutrient components of wild cocoyam Colocasia esculenta (L) Scott. Corm. Pakistan Journal of Nutrition 10(1):29-34.
Crossref
|
|
|
|
|
Oluduro AO, Bakare MK, Omoboye OO, Dada CA, Olatunji CI (2011). Antibacterial effect of extracts of Acalypha wilkesiana on gastrointestinal tract pathogens and bacteria causing skin infections in neonates. Ife Journal of Science 13:2.
|
|
|
|
|
Onah JO, Akabue PI, Okide JF (2002). The kinetics of reversal of pre-sickled erithrocytes by the aqueous extract of Cajanus cajan. Phytotherapy Research. 16(8):748-750.
Crossref
|
|
|
|
|
Onasanya A, Mignouna HD, Thottappilly G (2003). Genetic fingerprinting and phylogenetic diversity of isolates of Staphylococcus aureus from Nigeria. African Journal of Biotechnology 2:246-250.
Crossref
|
|
|
|
|
Oyewole OI, Malomo SO, Adebayo JO (2008). Comparative studies on antisickling properties of thiocyanate, tellurite and hydroxyurea. Pakistan Journal of Medical Sciences 24(1):18-22.
|
|
|
|
|
Pant J, Rai SK (2007). Occurrence of Staphyloccous aureus in Hospital Environment and Staffs in Teaching Hospital in Katmandu, Nepal. Nepal Association for Medical Laboratory Science 8:72-73.
|
|
|
|
|
Paul MO, Aderibie DA, Sule CZ (1982). Antimicrobial sensitivity patterns of hospital and non-hospital strains of Staphylococcus aureus isolated from nasal carriers. Journal of Hygiene 89:253-260.
Crossref
|
|
|
|
|
Singleton VL, Orthofer RL, Rosa M (1999). Analysis of total phenols and other oxidation substrates and antioxidants by means of folin-ciocalteu reagent. 299:152.
|
|
|
|
|
Sneath PH, Sokal RR. (1973). The Principle and Practice of Numerical Classification. In: Numerical Taxonomy, Kennedy, D. and R.B. Park (Eds.). Freeman, San Francisco.
|
|
|
|
|
Valgas C, De Souza ME, Smania FA (2007). Screening methods to determine antibacterial activity of natural products. Brazilian. Journal of Microbiology 38:369-380.
Crossref
|
|
|
|
|
Walson D (1989). Principles and Practice of Statistics in Biological Research, 2nd ed. ISBN 0-7167-1254-7, P 53.
|
|
|
|
|
Warren DK, Liao RS, Merz LR, Eveland M, Dunne WM Jr. (2004). Detection of methicillin-resistant Staphylococcus aureus directly from nasal swab specimens by a real-time PCR assay. Journal of Clinical Microbiology 42:5578-5581.
Crossref
|
|