ABSTRACT
Syringomycin E (SRE) is a phytotoxin produced by Pseudomonas syringae pv syringae with high potential as a safe and effective therapy for the control of human fungal infections and as a preservative for the food industry. In this study, 27 strains of P. syringae pv syringae were isolated from plums, potatoes, apricots and peaches, of which P. syringae pv syringae 120 (PS120) showed the highest SRE production levels. Furthermore, random mutagenesis induced by ultraviolet (UV) radiation resulted in the generation of a P. syringae pv syringae mutant that produced 30% more SRE than the parental strain PS120. To elucidate molecular mechanism underlying the higher SRE production ability of the mutant strain, syrB1 and syrB2 genes, which are known to be involved in SRE production, were cloned and sequenced. The nucleotide and amino acid sequences analysis showed that UV radiation induced numerous mutations at the AMP binding site on adenylation domain of syrB1, while no mutation was detected in syrB2 gene. Real-time polymerase chain reaction (PCR) results showed that expression of syrB1 gene in mutant strain was six-fold higher than that of PS120 strain. Taken together, these results suggest that the mutations at AMP binding site and overexpression of syrB1 were responsible for increased biosynthesis of SRE in P. syringae pv syringae.
Key words: Antifungal protein, syringomycin E, UV mutagenesis, Pseudomonas syringae pv syringae, syrB1.
The widespread resistance to commonly used antimicrobials and an increasing public health concern about the presence of chemicals in the food-chain have stimulated search for novel, effective and safe antimicrobial compounds. As a result, new resources and approaches are being explored to identify/develop the next generation of antimicrobials. Syringomycin has emerged as a new class of peptide presenting strong antimicrobial activity. For example, numerous studies have demonstrated the effectiveness of syringomycin E (SRE) against multiple filamentous fungal and yeast pathogens (Mazu et al., 2016; Im et al., 2003; Bender et al., 1999; De Lucca and Walsh, 1999; Fukuchi et al., 1992). The SRE can kill 95 to 99% of Aspergillus strains (Aspergillus flavus, Aspergillus niger, and Aspergillus fumigatus) and Fusarium (Fusarium moniliforme and Fusarium oxysporum) in concentrations from 1.9 to 7.8 μg/ml, but is non-toxic to mammalians and plant cells (De Lucca and Walsh, 1999; Takemoto et al., 2010). The SRE is also more effective at killing germinated spores of Aspergillus and Fusarium species than other peptides such as cecropin A, cecropin B and dermaseptin (De Lucca and Walsh, 1999). Kawasaki et al. (2016) showed that SRE inhibits 50 and 90% of P. ultimum oospore germination at 31.3 and 250 μg/ml, respectively.
In vitro growth inhibition experiments have further shown that SRE is toxic to a number of fungi such as A. fumigatus (opportunistic fungal pathogen) and Mucor species, Trichophyton species (parasitic skin fungus), responsible for causing fungal infections in humans (Muedi et al., 2011). In addition, the potential of SRE as an effective preservative for agricultural products has also been demonstrated; SRE was reported to be effective against a range of fungi involved in food spoilage including Penecillium digitatum in lychee, and Aspergillus, Rhizopus and Fusarium in orange, lychee, mango and dragon.
Syringomycin E, a natural phytotoxic, is a small cyclic lipodepsinonapeptide (ca. 1,200 Da) produced by the plant bacterium P. syringae pv. syringae (Guenzi et al., 1998; Raaijmakers et al., 2006). P. syringae pv. syringae also produces the large cyclic lipodepsipeptide, syringopeptins (Bensaci et al., 2011). The syringomycin gene cluster in P. syringae pv. syringae contains four genes involved in the biosynthesis of syringomycin E, including SyrB1, SyrB2, SyrC and SyrE (Guenzi et al., 1998); of which expression levels are regulated by salA, syrG, and syrF (Wang et al., 2006; Vaughn and Gross, 2016). SyrB1 and syrB2 function in generating one of the unusual amino acids of SRE scaffold, 4-Cl-L-Thr residues. SyrB1 gene with approximately 2 kb in length encodes for a 66 kDa enzyme consisting of an adenylation (A) domain which is responsible for amino acid selection and activation, and a thiolation (T) domain which is responsible for thioesterification of the activated substrate (Bender et al., 1999). The A domain catalyzes the activation and binding of L-Thr to phosphopantetheninyl in T domain, where it undergoes halogenation catalyzed by SyrB2 to create 4-Cl- thr-S- SyrB1 (Vaillancourt et al., 2005). Next, SyrC enzyme transfers the 4-Cl-Thr group from SyrBs1 to SyrE to create the complete SRE (Singh et al., 2007). Syringomycin is a cyclic lipodepsinonapeptide composed of a 3-hydroxy fatty acid tail. SRE, which contains a 3-hydroxy dodecanoic acid tail, is the major form produced by P. syringae pv.syringae strain B301D (Bender et al., 1999). Syringomycin is synthesized via a nonribosomal peptide synthetase system (Bender et al., 1999; Zhang et al., 1995). The syringomycin gene cluster is located in a 55 kb DNA region in the genome of B301D strain containing genes that are responsible for biosynthesis, regulation, and secretion of syringomycin (Lu et al., 2002; Quigley et al., 1993; Zhang et al., 1995, 1997).
The lower production efficiency of SRE by P. syringae pv. Syringae has been the major barrier to its cost-effective production and industrial application. Currently, much effort is focused on finding new ways to improve SRE production efficiency and develop SRE-based fungicides. Hence, the key objectives of this study was to determine if exposure to UV could be used to generate P. syringae pv. syringae mutants with enhanced SRE production efficiency and to examine molecular mechanisms, especially the role of syrB1 and syrB2 genes.
Bacteria isolation
P. syringae pv. syringae was isolated from stone fruit trees such as plums, apricots or peaches, and potatoes in Hanoi, Vietnam according to the method of Mohammadi et al., (2010). Briefly, biological samples were surface-sterilized by 70% ethanol for 2 min before incubation in distilled water with shaking for 2 h. 100 µl of suspension were plated on CMB agar supplemented with 5% sucrose. Colonies that produced fluorescence under UV light were picked and grown on King’s B media; gram stained and identified by standard biochemical assays for Pseudomonas species. Identification methods include catalase, oxidase, arginine dihydrolase, gelatin hydrolase and nitrate reductase assays together with levan production. Strains with correct biochemical activity were finally confirmed as P. syringae by 16S rDNA analysis.
Syringomycin production bioassay
The ability to produce SRE of P. syringae pv syringae was determined as described previously (DeVay and Gross, 1977). In brief, bacterial cells were grown overnight in nutrient broth yeast (NBY) medium at 25°C. The cells were harvested by centrifugation, washed with sterile deionized water, and adjusted to a final cell density of ∼2 × 108 CFU/ml. Five microliters of the bacterial suspension was inoculated to the previously prepared PDA plates
and incubated at 25°C for 4 days. The inoculated plates were overlaid with Geotrichum candidum VTCC-Y-0483 as an indicator organism and were further incubated for 24 h at 25°C in the dark. Clear zones of fungi growth inhibition around bacterial colonies were considered as an indicator of syringomycin production. One unit of antifungal activity was defined as the amount of the final dilution of cell-free extract in 1 mM HCl required to completely inhibit the growth of G. candidum in the area where a 10 µl droplet was applied on PDA plate (Sinden et al., 1971). Each assay was repeated independently three times with three plates per replicate.
The presence of syringomycin in P. syringae fermentation broth was confirmed by HPLC and LC-MS as previously described (Singh et al., 2007). Briefly, cells from 24-h cultures in NBY media at 25°C were pelleted by centrifugation, and supernatant extracted with water-saturated n- butanol at pH 2 before purification by cation-exchance chromotography and analysis by HPLC. HPLC was carried out with 20 µl of sample loaded onto a octadecylsilane bounded silica (5 µm) column (25 cm × 4.6 mm). The mobile phase, a mixture of 50 volume of 0.2% (w/v) acetic acid in water, 25 volume of acetonitrilc and 25 volume of methanol, was filtered and de-gased. HPLC was performed at a flow rate of 1 ml/min and observed at 240 nm. LC-MS analysis was performed on an LCMS-QP8000 a spectrometer (Shimadzu, Japan) with a Vydac C18 LC-MS column (Thermo Fisher Scientific, USA).
Random mutagenesis
P. syringae pv syringae parental strain was cultured at 180 rpm at 26°C on nutrient broth (NB) medium for 16 h. Biomass was collected by centrifugation; pellet was re-suspended in sterile distilled water to an OD of 0.5 to 0.6 and subjected to a UV light treatment with power of 40 W at various time doses. After 48 h culturing at 26°C, cells were plated to count the surviving colonies.
DNA preparation
P. syringae pv syringae mutant and parental strains were grown for 48 h at 28°C in SRM medium and collected by centrifugation at 16128 rpm for 2 min. Total DNA extraction was performed using PureLinkTM Genomic DNA Mini Kit (Invitrogen, USA) according to the manufacturer’s instructions. DNA was stored at -20°C until used.
Cloning and sequencing of 16S rRNA, syrB1 and syrB2 genes
The 16S rDNA, syrB1 and syrB2 genes were amplified by PCR with the specific primer pairs (Table 1) using Thermo cycler engine (AB system). The reaction mixture (25 µl) containing 12.5 µl of master mix (Thermo Fisher Scientific, USA) 10 pmol of each forward and reverse primers and 2 µl total DNA. The PCR conditions were as follows: 94°C for 3 min, followed by 40 cycles of 94°C for 30 s, 56°C for 40 s, 72°C for 1 min 30 s, and a final extension at 72°C for 7 min samples were then stored at 4°C until used. The PCR products were purified using the GENCLEANÒ II Kit (Q-BIO gene, Carlsbad, CA, USA), and directly cloned into a TOPO TA cloning vector (Invitrogen, USA) according to the manufacturer’s instructions and sequenced with ABI PRISMÒ 3100 Genetic Analyzer (Applied Biosystems, USA) using M13 forward primer and M13 reverse primer.
Real-time RT-PCR assay
Total cellular RNA was extracted using the TRIzol reagent (Invitrogen, USA) as per the manufacturer’s instructions. The extracted RNA was treated with DNase I at 37°C for 3 h to remove DNA. Reverse transcription was carried out with the Superscript TM III First-Strand Synthesis System (Invitrogen, USA) according to the manufacturer’s instructions. In detail, 500 ng mRNA was used as template for reverse transcription reaction. The cDNA was then used as a template for qualitative real-time PCR analysis using specific primers (Table 2). As a control for the input RNA, level of a housekeeping gene 16S rRNA was also assayed. The data was analyzed by LightCycler 2.0 software (Roche, USA).
Screening of P. syringae strains for SRE production
In this study, a number of bacterial strains were isolated from leaf tissues of plums, peaches, apricots, tomatoes and screened for production of syringomycin as described earlier. Among them, 120 strains were P. syringae pv syringae strains as indicated by biochemical assays and 16S rDNA analysis (data not shown). 14 strains produced antifungal metabolites characteristics of syringomycin and PS120 strain showed the highest amount of antifungal activity (36 U/ml) (Table 3).
Characterization of antifungal compounds produced by strain PS120
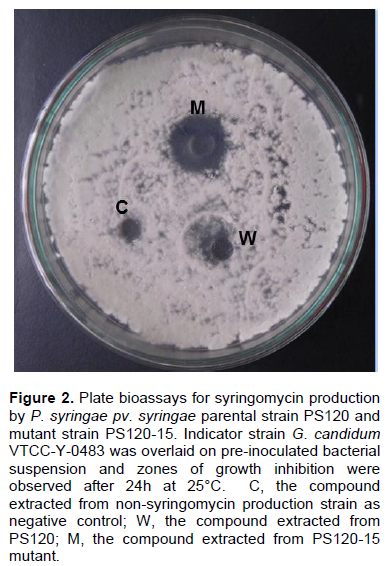
The antifungal compounds of PS120 were extracted and purified and analyzed by SDS-PAGE; similarly extracted metabolites from a non-syringomycin producing strain were used as a negative control. Fifteen micrograms of the purified compound and an equal amount of standard SRE (Sigma, USA) were separated on 12% polyacrylamide gel, and then stained with Coomassie-blue. The results showed that the purified compound was a protein with MW similar to that of the standard SRE (Figure 1). To determine whether this compound was syringomycin, its antifungal activity was examined against G. candidum strain VTCC-Y-0483 using a standard plate bioassay. It was observed that this compound produced large zone of inhibition against G. candidum. In contrast, no zone of inhibition was observed with the compound extracted from non-syringomycin producing strain (Figure 2).
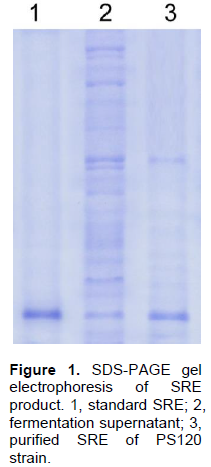
Further analysis of the purified compound by HPLC revealed that it has the same retention time as the standard SRE (Sigma, USA). The standard SRE appeared at 4.283 min, while that from P. syringae pv syringae PS120 was at 4.281 min (Figure 3), indicating the purified compound was SRE. The identity of the purified compound was further analysed using ESI mass spectroscopic analyses. It was found that the purified compound generated MH+ species with molecular weights of 1224 to 1226 and major fragmentation species with molecular weight of 613 and 614 (Figure 4) that are consistent with the proportion of 2 isoforms of chlorine (35Cl and 37Cl), the key characteristics of SRE.
Enhancing the production level of SRE
PS120 strain was exposed to various doses of UV to create mutant strains with enhanced SRE production. Following initial screening, 334 mutants were selected that survived after exposure to UV light for 180 s. The SRE production capacity of these mutants was assessed from their ability to inhibit the growth of G. candidum. The mutants with increased SRE production were selected following the method described by DeVay and Gross (1977). Of the 334 mutants, five were found to produce significantly higher levels of SRE (20 to 32.2%) compared with the parental PS120 strain (Figure 5). P. syringae mutant (PS120-15) showed the highest SRE production (47.6 U/ml), that is 32.2% higher than the parental strain (36 U/ml). This result was further supported by a larger inhibitory zone to G. candidum induced by PS120-15 compared to PS120 (Figure 2). PS120-15 mutant was further studied to understand the molecular mechanism responsible for the increased SRE production.
Analysis of syrB1 and syrB2 genes and their expression
SyrB1 and syrB2 genes from both wild type (PS120) and mutant (PS120-15) strains were PCR amplified using specific primers as described earlier. The PCR products were then cloned, sequenced and analyzed using Bioedit (Tom Hall, Ibis Biosciences). While no change in syrB2 sequence was detected following UV treatment, an additional nucleotide fragment was found inserted in syrB1 sequence in PS120-15 mutant that led to the insertion of an amino acid fragment in syrB1 protein (Figure 6). Further analysis revealed that the inserted fragment was located at the AMP binding site (between the position of 304-408 of syrB1), an important element in the biosynthesis of SRE (Kleinkauf and Von Dohren, 1996).
The expression of syrB1 and syrB2 in wildtype and mutant was then measured by real-time PCR assay. The data showed that expression level of syrB1 in mutant strain was six-fold higher than that in the wild type strain (Figure 7). On the other hand, the expression level of syrB2 remained unaffected (data not shown). This suggests that the insertion of amino acid fragment in syrB1 was associated with increased expression of syrB1 and that this may be responsible for increased production of SRE in the mutant strains.
Because of the SRE antimicrobial activities and due to the fact that it has no impacts on both human health and environment, SRE has been the focus of much research with ultimate aim to develop new generation of antimicrobials. SRE has been shown to be an effective fruit preservative agent against a range of fungal species including G. candidum, Rhodotorula pilimance, Botrytis cinerea, Fusarium, Pythium ultimum, Rhizoctonia, Greeneria uvicola, Aspergillus japonicus, Mucor species, Trichophyton species, Curvularia brachyspora, Nigrospora sphaerica, Penicillium thomii and Penicillium sclerotiorum (Breen et al., 2015; D'aes et al., 2010; Bull et al., 1998; Janisiewicz and Bors, 1995). In addition, it exhibits toxicity against Hela cells at 20-times higher concentrations than its fungicidal concentration that kills 95% of pathogenic fungi (De Lucca et al., 1999).
A number of studies have been conducted to isolate P. syringae pv. Syringae strains which produce SRE (Janisiewicz and Bors, 1995; Cirvilleri et al., 2005; Singh et al., 2007). In this study, a total of 260 P. syringae pv syringae strains was isolated and 27 strains produced antifungal metabolites characteristics of syringomycin was selected. Among these, PS120 showed the highest amount (36 U/ml). However, the production efficiency of SRE of PS120 is still low and has been the major barrier to its cost-effective production and industrial application. With the aim to create P. syringae pv. syringae mutants with enhanced ability to produce SRE, random muta-genesis approach was applied using UV light treatment. The effectiveness of UV light induced mutagenesis has previously been reported (Ikehata and Ono, 2011), such as to increase production of clavulanic acid by Streptomyces clavuligerus (Lee et al., 2002) and bio-insecticides production in Bacillus thuringenesis (Ghribi et al., 2004). Interestingly, five of the 334 mutants, resulting from random mutagenesis induced by exposure of PS120 strain to UV light, produced significantly higher levels (20 to 32%) of SRE than the parental strain PS120. Out of the 334 mutants screened, a mutant strain (PS120-15) producing the highest level of SRE was selected that synthesized SRE up to 30% higher than its parental strain. These findings are consistent with the results of earlier studies that used UV light-induced mutagenesis to enhance production of bioinsecticides by B. thuringiensis (Ghribi et al., 2004) and clavulanic acid by S. clavuligerus (Muedi et al., 2011).
Although significant efforts have been made toward understanding the molecular mechanisms that trigger and control the biosynthesis of SRE, much is still unknown about this process (Roongsawang et al., 2011). Transposon mutagenesis performed by Xu and Gross (1988) and Zhang and Gross (1995) has proven that a chromosomal region larger than 25 kb is involved in the biosynthesis of SRE. Within this region, four genes, namely, syrB, syrC, syrD and syrP have been sequenced and partially characterized (Martinie et al., 2015; Singh et al., 2007; Gross, 1991; Zhang and Gross, 1997; Quigley et al., 1993). SyrD has similar sequence to the ATP binding cassette transporter superfamily, thus, it is hypothesized that syrD product is involved in the transportation of SRE across the cytoplasmic membrane (Quigley et al., 1993). Further sequencing investigation by Guenzi et al. (1998) showed that syrB is actually organized in two ORFs, syrB1 and syrB2 encoding a 67 kD protein and a 36 kD protein, respectively. The 67 kD protein encoded by syrB1 carries all known conserved regions of the amino acid binding modules of peptide synthetases.
It is well documented that syrB1 and syrB2 are involved in the biosynthesis of SRE of P. syringae pv syringae. It was hypothesized that UV radiation is likely to induce mutations and/or enhance the expression of these two enzymes to influence the production of SRE directly or indirectly. To test this hypothesis, syrB1 and syrB2 genes was cloned and sequenced. Previous studies have shown while syrB1 carries all consensus sequences of amino acid binding module of peptide synthetases, syrB2 sequence shows little to no consensus to other known peptide synthetase (Guenzi et al., 1998). In this study, the sequence analysis showed that an insertion occurred at AMP binding site in syrB1 gene at nucleotide position 304 to 408, which belongs to adenylation domain responsible for recognizing and activating a specific amino acid (Kleinkauf and Von Dohren, 1996; Marahiel et al., 1997). Previous study has indicated that SRE is synthesized by nonribosomal mechanism (Singh et al., 2007) with the participation of syrB1, syrB2, syrC and syrE. Nonribosomal peptide synthetases, such as syrB1 and syrB2, are multimodular enzymes that produce peptide via thiotemplate mechanism independent of ribosomes. Well-known products of nonribosomal peptide synthetases are antibiotics (such as penicillin, erythromycin, vancomycin), immune suppressants (such as cyclosporine and rapamycin) and antitumor agents (Lee et al., 2005). Minimal nonribosomal peptide synthetase module consists of an AMP-binding adenylation domain (A) for substrate recognition and activation; and a thiolation (T, also called peptidyl carrier protein) domain, downstream of A domain, functioning as cofactor binding site (Schwarzer and Marahiel, 2001). This A domain is highly conserved; therefore, mutations in AMP-binding site might lead to the improvements in SRE production ability of the mutant strain.
To test whether the UV treatment induced the expression of syrB1, a realtime PCR assay was conducted. The real-time PCR result showed that expression level of syrB1 in mutant strain was six-times higher compared with the wildtype strain. The increased expression level of syrB1 might be due to some acquired mutations in its promoter region and responsible for the increased biosynthesis of SRE. It is likely that insertion of new fragment of amino acid into the adenylation domain of SyrB1 increases its amino acid activation rate, indirectly boosting SRE biosynthesis. At present, the molecular mechanism(s) underlying the higher SRE production ability of the mutant strain was not fully understood. This study showed a successful enhance-ment of SRE production in P. syringae pv syringae using UV induced mutagenesis, proving involvement of at least one gene, syrB1. These results shed more lights into the biosynthesis pathway of SRE in P. syringae, giving directions for further characterization of the pathway in the future.
The authors declare that there is no conflict of interest.
REFERENCES
|
Bender CL, Alarcón-Chaidez F, Gross DC (1999). Pseudomonas syringae Phytotoxins: Mode of Action, Regulation, and Biosynthesis by Peptide and Polyketide Synthetases. Microbiol. Mol. Biol. Rev. 63:266-292.
|
|
|
|
Bensaci MF, Gurnev PA, Bezrukov SM, Takemoto JY (2011). Fungicidal Activities and Mechanisms of Action of Pseudomonas syringae pv. syringae Lipodepsipeptide Syringopeptins 22A and 25A. Front. Microbiol. 2:216
Crossref
|
|
|
|
|
Breen S, Solomon PS, Bedon F, Vincent D (2015). Surveying the potential of secreted antimicrobial peptides to enhance plant disease resistance. Front. Plant Sci. 6:900.
Crossref
|
|
|
|
|
Bull CT, Wadsworth ML, Sorensen KM, Takemoto JY, Austin RK, Smilanick JL (1998). Syringomycin E produced by biological control agents controls green mold on lemons. BioI. Control 12:89-95.
Crossref
|
|
|
|
|
Cirvilleri G, Bonaccorsi A, Scuderi G, Scortichini M (2005). Potential Biological Control Activity and Genetic Diversity of Pseudomonas syringae pv. syringae Strains. J. Phytopathol. 153:654-666.
Crossref
|
|
|
|
|
DeVay JE, Gross DC (1977). Production and purification of syringomycin, a phytotoxin produced by Pseudomonas syringae. Physiol. Plant Pathol. 11:13-28.
Crossref
|
|
|
|
|
D'aes J, De Maeyer K, Pauwelyn E, Höfte M (2010). Biosurfactants in plant-Pseudomonas interactions and their importance to biocontrol. Environ. Microbiol. Rep. 2:359-372.
Crossref
|
|
|
|
|
De Lucca AJ, Walsh TJ (1999). Antifungal peptides: novel therapeutic compounds against emerging pathogens. Antimicrob. Agents Chemother. 43:1-11.
|
|
|
|
|
De Lucca AJ, Jacks TJ, Takemoto J, Vinyard B, Peter J, Navarro E, and Walsh TJ (1999). Fungal lethality, binding, and cytotoxicity of syringomycin-E. Antimicrob. Agents Chemother. 43:371-373.
|
|
|
|
|
Fukuchi N, Isogai A, Nakayama J, Takayama S, Yamashita S, Suyama K, Takemoto JY, Suzuki A (1992). Structure and stereochemistry of three phytotoxins, syringomycin, syringotoxin and syringostatin, produced by pseudomonas syringae pv. syringae. J. Chem. Soc. Perkin Trans. 1. 1992(9):1149-1157.
Crossref
|
|
|
|
|
Ghribi D, Zouari N, Jaoua S (2004). Improvement of bioinsecticides production through mutagenesis of Bacillus thuringiensis by u.v. and nitrous acid affecting metabolic pathways and/or delta-endotoxin synthesis. J. Appl. Microbiol. 97:338-346.
Crossref
|
|
|
|
|
Gross DC (1991). Molecular and genetic analysis of toxin production by pathovars of Pseudomonas syringae. Annu. Rev. Phytopathol. 29:247-278.
Crossref
|
|
|
|
|
Guenzi E, Galli G, Grgurina I, Gross DC, Grandi G (1998). Characterization of the syringomycin synthetase gene cluster. A link between prokaryotic and eukaryotic peptide synthetases. J. Biol. Chem. 273:32857-32863.
Crossref
|
|
|
|
|
Ikehata H, Ono T (2011). The mechanisms of UV mutagenesis. J. Radiat. Res. 52(2):115-125.
Crossref
|
|
|
|
|
Im YJ, Idkowiak-Baldys J, Thevissen K, Cammue BP, Takemoto JY (2003). IPT1-independent sphingolipid biosynthesis and yeast inhibition by syringomycin E and plant defensin DmAMP1. FEMS Microbiol Lett. 223(2):199-203.
Crossref
|
|
|
|
|
Janisiewicz WJ, Bors B (1995). Development of a microbial community of bacterial and yeast antagonists to control wound-invading postharvest pathogens of fruits. Appl. Environ. Microbiol. 61:3261-3267.
|
|
|
|
|
Kleinkauf H, Von Dohren H (1996). A nonribosomal system of peptide biosynthesis. Eur. J. Biochem. 236:335-351.
Crossref
|
|
|
|
|
Lee BN, Kroken S, Chou DY, Robbertse B, Yoder OC, and Turgeon BG (2005). Functional analysis of all nonribosomal peptide synthetases in Cochliobolus heterostrophus reveals a factor, NPS6, involved in virulence and resistance to oxidative stress. Eukaryot. Cell 4:545-555.
Crossref
|
|
|
|
|
Lee SD, Park SW, Oh KK, Hong SI, and Kim SW. 2002. Improvement for the production of clavulanic acid by mutant Streptomyces clavuligerus. Letters in applied microbiology. 34: 370-375.
Crossref
|
|
|
|
|
Lu SE, Scholz-Schroeder BK, Gross DC (2002). Characterization of the salA, syrF, and syrG regulatory genes located at the right border of the syringomycin gene cluster of Pseudomonas syringae pv. syringae. Mol. Plant Microbe Interact.15:43-53.
Crossref
|
|
|
|
|
Marahiel MA, Stachelhaus T, Mootz HD (1997). Modular Peptide Synthetases Involved in Nonribosomal Peptide Synthesis. Chem. Rev. 97:2651-2674.
Crossref
|
|
|
|
|
Martinie RJ, Livada J, Chang WC, Green MT, Krebs C, Bollinger JM Jr, Silakov A (2015). Experimental Correlation of Substrate Position with Reaction Outcome in the Aliphatic Halogenase, SyrB2. J. Am. Chem. Soc. 137(21):6912-6919.
Crossref
|
|
|
|
|
Mazu TK, Bricker BA, Flores-Rozas H, Ablordeppey SY (2016). The Mechanistic Targets of Antifungal Agents: An Overview. Mini Rev. Med. Chem. 16(7):555-578.
Crossref
|
|
|
|
|
Mohammadi M, Ghasemi A, Rahimian H (2010). Phenotypic Characterization of Iranian Strains of Pseudomonas syringae pv. syringae van Hall, the Causal Agent of Bacterial Canker Disease of Stone Fruit Trees. J. Agric. Sci. Technol. 3:51-65.
|
|
|
|
|
Muedi HT, Fourie D, Mclaren NW (2011). Characterisation of bacterial brown spot pathogen from dry bean production areas of South Africa. Afr. Crop Sci. J. 19:357-367.
|
|
|
|
|
Quigley NB, Mo YY, Gross DC (1993). SyrD is required for syringomycin production by Pseudomonas syringae pathovar syringae and is related to a family of ATP-binding secretion proteins. Mol. Microbiol. 9:787-801.
Crossref
|
|
|
|
|
Raaijmakers JM, de Bruijn I, de Kock MJ (2006). Cyclic lipopeptide production by plant-associated Pseudomonas spp.: diversity, activity, biosynthesis, and regulation. Mol. Plant Microbe Interact. 19:699-710.
Crossref
|
|
|
|
|
Roongsawang N, Washio K, Morikawa M (2011). Diversity of Nonribosomal Peptide Synthetases Involved in the Biosynthesis of Lipopeptide Biosurfactants. Int. J. Mol. Sci. 12(1):141-172.
Crossref
|
|
|
|
|
Schwarzer D, Marahiel MA (2001). Multimodular biocatalysts for natural product assembly. Naturwissenschaften 88:93-101.
Crossref
|
|
|
|
|
Singh GM, Vaillancourt FH, Yin J, Walsh CT (2007). Characterization of SyrC, an aminoacyltransferase shuttling threonyl and chlorothreonyl residues in the syringomycin biosynthetic assembly line. Chem. Biol. 14:31-40.
Crossref
|
|
|
|
|
Takemoto JY, Bensaci M, De Lucca AJ, Cleveland TE, Gandhi NR, Palmer Skebba V (2010). Inhibition of Fungi from Diseased Grape by Syringomycin E-Rhamnolipid Mixture. Am. J. Enol. Viticulture 61:120-124.
|
|
|
|
|
Vaillancourt FH, Yin J, Walsh CT (2005). SyrB2 in syringomycin E biosynthesis is a nonheme FeII alpha-ketoglutarate- and O2-dependent halogenase. Proc. Natil. Acad. Sci. USA. 102:10111-10116.
Crossref
|
|
|
|
|
Vaughn VL, Gross DC (2016). Characterization of salA, syrF, and syrG Genes and Attendant Regulatory Networks Involved in Plant Pathogenesis by Pseudomonas syringae pv. syringae B728a. PLoS One 11(3):e0150234
Crossref
|
|
|
|
|
Wang N, Lu SE, Records AR, Gross DC (2006). Characterization of the transcriptional activators SalA and SyrF, Which are required for syringomycin and syringopeptin production by Pseudomonas syringae pv. syringae. J. Bacteriol. 188(9):3290-3298.
Crossref
|
|
|
|
|
Xu GW, Gross DC (1988). Evaluation of the Role of Syringomycin in Plant Pathogenesis by Using Tn5 Mutants of Pseudomonas syringae pv. syringae Defective in Syringomycin Production. Appl. Environ. Microbiol. 54:1345-1353.
|
|
|
|
|
Zhang JH, Quigley NB, Gross DC (1995). Analysis of the syrB and syrC genes of Pseudomonas syringae pv. syringae indicates that syringomycin is synthesized by a thiotemplate mechanism. J. Bacteriol. 177:4009-4020.
Crossref
|
|
|
|
|
Zhang JH, Quigley NB, Gross DC (1997). Analysis of the syrP gene, which regulates syringomycin synthesis by Pseudomonas syringae pv. syringae. Appl. Environ. Microbiol. 63:2771-2778.
|
|