ABSTRACT
Acid phosphatase from mung bean (Vigna radiata) germinated seeds was partially purified via ammonium sulfate precipitation and fully purified via gel excision and heat denaturation, yielding a 29 kDa protein. Using 4-nitrophenylphosphate as a substrate, Vmax and Km values of 0.10 µmol/min and 0.27 mM, respectively, were obtained. The acid phosphatase was heat resistant, enhanced by the presence of Fe2+ and Mn2+ salts, and inhibited by the presence of citrate. Mung bean acid phosphatase reacted immunologically with primary antibodies against Solanum tuberosum acid phosphatase, while polymerase chain reaction (PCR) analyses suggest there are some common sequences between mung bean acid phosphatase DNA and that of Arabidopsis thaliana acid phosphatase vegetative storage protein. This suggests plant acid phosphatases are structurally as well as functionally related.
Key words: Acid phosphatase, mung bean, Vigna radiata, vegetative storage protein.
Phosphatases (3.1.3) are a diverse group of enzymes found in most living systems. Historically, they have been less well studied than protein kinases, but recently there has been more interest in both protein and genetic characterization and regulation (Bheri et al., 2021; Schweighofer and Meskiene, 2015; Brautigan, 2013; Uhrig et al., 2013; Anand and Srivastava, 2012; Moorman and Brayton, 2021). In plants, phosphatases appear to have the important roles of liberating and securing phosphates for growth, especially in germinating seeds (Duff et al., 1994; Anand and Srivastava, 2012). These enzymes are abundant and robust enough that their extraction and analyses are routinely used as a laboratory experiment in undergraduate biochemistry courses (Moorman and Brayton, 2021) as done in Northern Michigan University. Acid phosphatases, which are optimally active at acidic pH, and are the focus of this paper, often appear to catalyze reactions with a broad spectrum of substrates, such as 4-nitrophenyl phosphate, ATP, pyridoxal-5’-phosphate, creatine phosphate, etc. (Verjee, 1969; Joyce and Grisolia, 1960). Several subclasses of acid phosphatase (AP) enzymes include purple acid phosphatases (PAP) which contain a chromogenic iron center (Anand and Srivastava, 2012; Tran et al., 2010; Schenk et al., 2000), tartarate-resistant acid phosphatases which are not affected by tartrate, vegetative storage proteins (VSP) which, in addition to phosphatase activity, also store essential amino acids for growing plants (Staswick, 1994), and DXDXT phosphatases which all share the aspartate, aspartate, and threonine consensus sequence (Collet et al., 1998). Some of these DXDXT phosphatases also have other consensus sequences such as GDXXXD (Thaller et al., 1998); clearly aspartate is an important amino acid for these enzymes. Arabidopsis thaliana (thale cress) is a model organism for plants; this means its genome is well-studied and it is often used as a blueprint for plant studies. According to genetic analyses of the A. thaliana, putative PAP are the most abundant AP (The Arabidopsis Genome Initiative, 2000; Li et al., 2002), while the actual number of AP enzymes present in a species is related to its polyploidy. A number of studies have been done on plants deprived of phosphate and AP expression increases during times of low phosphate, but the expression varies among plant tissue type, that is, roots vs. leaves (Li et al., 2002; Mission et al., 2005; Wu et al., 2003; Bargaz et al., 2013; Baldwin et al., 2001; Tejera Garcia et al., 2004).
Vegetative storage proteins (VSPs) are thought to be used by plants as a depot for essential amino acids and are metabolized when the need arises, such as during times of growth and injury (Staswick, 1994). At least one of the A. thaliana VSPs, VSP2, has been shown to have not only phosphatase activity at acidic pH, but also functions as an anti-insect protein (Liu et al., 2005).
Furthermore, the anti-insect activity, or defense role, is tied to the phosphatase activity as deletion of a critical aspartate in the DXDXT consensus sequence results in loss of both activities. To date sequence analysis of the A. thaliana genome suggests over 40 putative AP proteins with no specific functions assigned (The Arabidopsis Genome Initiative, 2000; Li et al., 2002), therefore much remains to be sorted out for this interesting and diverse group of enzymes.
A number of years have been spent using acid phosphatase from Vigna radiata (mung bean) sprouts to examine kinetics and protein purification. It is simple to germinate mung bean seeds and the AP activity is easily assayed with 4-nitrophenylphosphate. However, in our hands, the enzyme does not purify with ion exchange chromatography. This is in contrast to the many literature protocols for isolating AP from plants such as common bean (Tejera Garcia et al., 2004), wheat germ (Waymack and Van Etten, 1991), and yellow lupin (Olczak et al., 1997) using anion exchange resins, while protocols for AP isolation from other plants such as soybean (Hegeman and Grabau, 2001), and peanut (Gonnety et al., 2006) employ both anion and cation exchange resins over multiple steps. Nadir et al. (2012) reported using anion exchange chromatography in their partial purification of a 29 kD acid phosphatase protein from germinated mung bean seeds (Nadir et al., 2012), while in 2014 this same research group described the purification of another 29 kD acid phosphatase from the leaves of germinating mung bean seeds using cation exchange chromatography (Saeed et al., 2014). Clearly these reports and numerous isolation procedures suggest that there are several AP isoenzymes in plants and purification is not straight-forward. Surchandra et al. (2012) published a report that described an essentially one-step purification of mung bean AP with ammonium sulfate precipitation; however, after several years of trying, these results could not be duplicated. The current paper describes two methods of isolating mung bean AP, (1) using heat denaturation and (2) using band excision from an SDS polyacrylamide gel. A partial gene was also isolated for mung bean AP using DNA primers based on the sequence of an A. thaliana VSP2 acid phosphatase and compared its expression at two different temperatures.
Chemicals and reagents
All chemicals and reagents, unless otherwise noted, were of research grade and obtained from commercial chemical vendors.
Growth of seedlings
Organic mung bean seeds were obtained from a local food cooperative. The seeds were soaked overnight in water, drained, spread out on paper towels, and covered with aluminium foil to ensure darkness for germination. The seeds were allowed to germinate at room temperature or at 37°C for an average of four days.
Extraction of protein from germinated seedlings
Sprouted seeds (20 to 10 g) were either chopped in a small food processor or mashed thoroughly with a mortar and pestle, and then mixed with cold 0.1 M sodium acetate, pH 5.2 (2.5 mL extraction buffer per gram seed mass). After filtering through cheesecloth the resulting filtrate was centrifuged for 30 min at 12,000 x g, 4°C. The pellet was discarded and the supernatant labeled as “the crude extract”.
Ammonium sulfate precipitation of crude extract
The crude extract was made about 30% in (NH4)2SO4 (0.166 g/mL) and incubated on ice for 30 min. After centrifuging for 30 min at 12,000×g, 4°C, the resulting supernatant was brought to about 70% in (NH4)2SO4 (0.253 g/mL) and incubated for an additional 30 min on ice. The final centrifugation was for 30 min at 17,400×g, 4°C. The pellet was resuspended in 2 mL of cold 0.1 M sodium acetate, pH 5.2 and dialyzed against the same over two days at 4°C in a total of 8 L with one exchange after 24 h.
Assay of acid phosphatase activity
The activity of acid phosphatase was measured using 4-nitrophenylphosphate (4NPP). Assays were conducted at room temperature with 0.1 to 12 mM of substrate, 0.1 M sodium acetate, pH 5.2, buffer in a final volume of 3.0 mL. Termination of the enzyme activity was done with NaOH (0.1 mL of 8 M yielding a final concentration of 0.27 M). An aliquot of enzyme (usually 0.1 mL or 0.05 mL) was used to initiate the reaction and it was terminated either at 3.0 or 10.0 min with NaOH. Concentration of the product, 4-nitrophenol, was measured spectrophotometrically at 410 nm. The spectrophotometer was blanked for each concentration of PNPP with the same assay concentrations and conditions except that water was used in place of the enzyme. Rates were calculated in μmol/min using 19.3 mM-1cm-1 as the molar absorptivity of 4-nitrophenol determined with pure 4-nitrophenol in 0.1 M sodium acetate, pH 5.2.
Determination of Km and Vmax
Maximum velocity (Vmax) and the Michaelis-Menten constant (Km) were calculated over a range of substrate from 0.1 to 12 mM for a 10 min assay. Data were plotted and curve-fitted with Origin graphing software using the Hill function, Vmax(Xn/Kn + Xn) where n = number of binding sites and was set to 1. The data were also analyzed linearly in Lineweaver-Burk, Eadie-Hofstee and Hanes-Woolf plots (Copeland, 2000) and parameters averaged.
SDS- PAGE analysis
Samples were run under denaturing conditions (2-mercaptoethanol) on BioRad 4-20% tris-glycine mini-gels (Laemmli, 1970). Running buffer contained tris, glycine, and SDS; gels were run at 150 V for about 1 h. All gels were stained with 0.1% w/v Coomassie Blue R250 in 50% methanol, 7.5% acetic acid, and 42.5% water, and destained with methanol/acetic acid/water in a 25:10:65 ratio.
Band excision of AP protein from SDS polyacrylamide gel
Samples were run under non-denaturing (no 2-metcaptoethanol) conditions on a BioRad 4 to 20% tris-glycine mini-gel. All of the individual wells were removed from the gel and it was run as one large well in which 160 μL of protein sample plus non-reducing tracking dye were added. The gel was run at 150 V for about 1 h and not stained. The bands (horizontal bands running the width of the gel) of the partially-purified acid phosphatase were cut out of the gel with a clean razor blade and crushed up in 0.1 M sodium acetate, pH 5. The gel fragments were incubated in the sodium acetate buffer overnight at 4°C, physically removed from the buffer the following day. The buffer was concentrated and exchanged with fresh 0.1 M sodium acetate, pH 5.0 using Amicon 10 centrifuge filters.
Heat denaturation studies
Dialyzed mung bean protein was heated to various temperatures for 15 to 30 min. Samples were consequently centrifuged at 10,000×g in a microcentrifuge for 3 to 5 min to remove precipitate.
Western blot analysis
Gels were run as described earlier under non-denaturing (no 2-mercaptoethanol). Instead of staining and fixing gels, they were subjected to electrical transfer in 20 mM tris, 192 mM glycine, and 20% (v/v) methanol at 4°C overnight. Antibodies used were either a primary rabbit antibody against Solanum tuberosum (potato) acid phosphatase (Agrisera) followed by a goat anti-rabbit peroxidase conjugate secondary (Rockland), or a rabbit anti-potato acid phosphatase peroxidase conjugate (Rockland) that did not require a secondary antibody. All antibodies were used at vendor recommended dilutions. Incubation buffer was a tris-buffered saline that contained 2% (w/v) instant dry milk, and 0.05 to 0.1% (v/v) tween 20.
Agarose gel analysis
All DNA analyses were performed on 1to 2% (w/v) agarose gels with tris-borate-EDTA running buffer at 80 to 150 V for 1 h (Davis et al., 1994). Detection was with ethidium bromide.
DNA extraction from mung bean sprouts
Mung bean sprouts were germinated at room temperature as specified earlier, frozen in liquid N2, ground, and extracted with the Promega Wizard or the QIAGEN Genomic DNA Purification kits using the plant protocol. Purity and size of DNA was checked by agarose gel electrophoresis.
Polymerase chain reaction (PCR) of mung bean DNA
Primer sets were designed to amplify a region of chromosome 5 of A. thaliana. The region of chromosome 5 corresponded to the VSP2 acid phosphatase gene (NCBI database accession# AB006778.1) and comprised 8,500275 to 8,500,296 bp for the forward primer and 8,502,218 to 8,502,242 bp for the reverse primer. The two primer sets, #1 and #2, differed only by an additional nucleotide at one end, and were designed by the primer choosing feature of the NCBI’s BLAST program.
Lyophilized primers (25 nmol) were obtained from Integrated DNA Technologies, Inc. The ribulose-1,5-bisphosphate carboxylase/oxygenase enzyme (Rubisco; rbcL) primer was obtained from the Carolina Biological Supply Company. PCR reactions were run using commercially available 10 mM nucleotide mix, 25 mM MgCl2, amplification grade water, Promega’s GoTaq polymerase (5U/μL) and green GoTaq buffer, and 6 pm of each primer. A Perkin Elmer 480 thermal cycler was used for amplification with 35 cycles of 94°C for 30 s, 54°C for 45 s, and 72°C for 45 s, after a hot start of 94°C for 5 min.
Reverse transcriptase procedure
Total RNA was isolated from mung bean seeds germinated at room temperature by crushing them in liquid nitrogen and following vendor protocols in the RNAqueous-4PCR total RNA Isolation Kit (Ambion/ThermoFisher Scientific). RNA was quantitated via UV spectroscopy and then subjected to reverse transcription using a GoScript Reverse Transcription System A5000 (Promega). The cDNA was then subjected to PCR as described earlier.
Purification of acid phosphatase
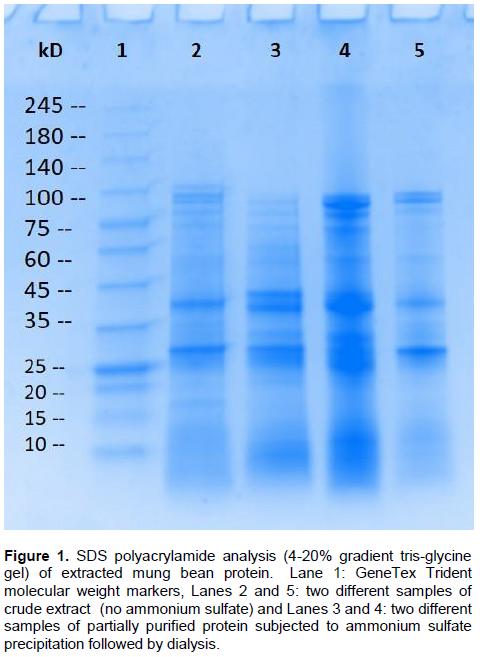
Following the protocol outlined in Surchandra et al. (2012), ammonium sulfate was used in a two-step process to partially purify acid phosphatase (AP) in order to precipitate and concentrate the protein. The Surchandra et al., (2012) process was to use a 30% cut with ammonium sulfate first followed by a 70% cut. A variety of ammonium sulfate concentrations and steps including single cuts of 50 to 80% ammonium sulfate and two-step cuts having final values of salt at 50 to 90% was also experimented with. It was found that an ammonium sulfate cut of 70% led to the best AP activity, but there was not a drastic difference between the other salt values in terms of activity or purity (data not shown). For purposes of keeping to the Surchandra et al. (2012) protocol for comparisons, most data reported in this paper involve the two step ammonium sulfate precipitation with a final ammonium sulfate cut of 70%. Figure 1 shows the SDS PAGE of ammonium sulfate precipitation followed by dialysis against buffer. Lanes 2 and 5 are two different samples of crude protein extract, while lanes 3 and 4 are two different preparations of extract that has been salted out followed by dialysis. Clearly there are a multitude of bands in the ammonium sulfate-precipitated protein (lanes 3 and 4) and the protein is not pure. In fact there is not much difference between the crude extract and the salted-out protein. This is in contrast to the gel photo reported by Surchandra et al., (2012) which showed a single band at approximately 34,500 Daltons after only ammonium sulfate precipitation.
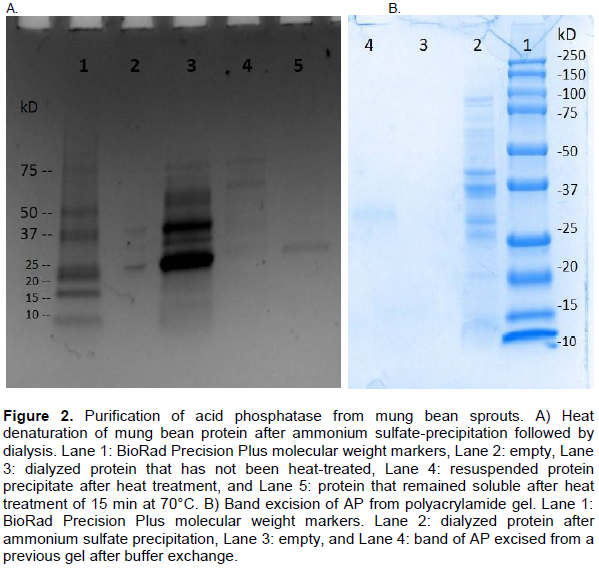
In order to fully purify the AP enzyme, we tried column chromatography. In our hands, the dialyzed AP did not purify further with cation or anion exchange, gel filtration, or lectin chromatography (data not shown). Heat denaturation followed by centrifugation did lead to a pure protein (Figure 2A, lane 5). These results suggest that mung bean AP is quite heat stable, it remained at least partially active up until 80°C (Figure 4). Bands from a gel of a partial purification in order to obtain pure protein were also excised. After buffer exchange to remove the SDS from the gel bands, it was found that the mung bean acid phosphatase molecular weight was about 29 kDa (Figure 2B). These results are similar to the estimated 34.5 kDa reported by Surchandra et al., (2012), and also fit the putative mung bean acid phosphatase sequence deposited in the NCBI protein database (Kang et al., 2014) which suggests a protein of 31 kDa. The literature reports of acid phosphatase isolation from plants describe either a low molecular weight enzyme of around 30 kDa (Zaman et al., 2020; Saeed et al., 2014; Nadir et al., 2012; Asaduzzaman et al., 2011; Zaman et al., 2020) or a higher molecular weight enzyme around 60 kDa (Anand and Srivastava, 2014; Xie and Shang, 2018). This study produced a low molecular weight AP enzyme. Both reducing and non-reducing gels were used and yielded the same results (data not shown) suggesting the low molecular weight AP is monomeric. It is unclear as to how Surchandra et al. (2012) managed to isolate a single protein with only an ammonium sulfate precipitation. We have one plausible idea; the gel analysis by the Surchandra group may have involved depleted Coomassie blue stain. In our hands any type of Coomassie blue stain has a limited shelf life; depleted stain does not adequately stain all proteins.
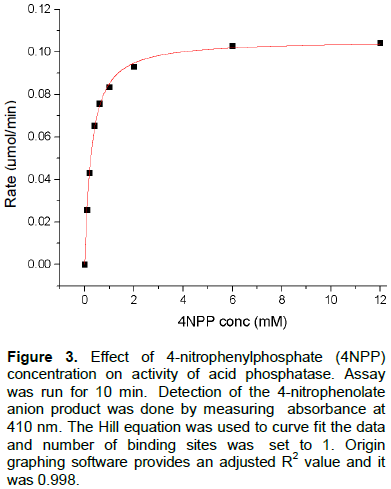
Activity assays with 4-nitrophenylphosphate
Because heat denaturation or excising protein from a gel led to either low yields or some significant loss of activity, many of the kinetic assays were performed with partially purified acid phosphatase. Plotting rate versus concentration of 4-nitrophenylphosphate substrate in a saturation plot (Figure 3) showed Michaelis-Menten type kinetics as saturation was achieved and the data fit to a rectangular hyperbola. The experiment was set up in a similar manner to Surchandra et al., (2012) so that we could directly compare the kinetic parameters maximum velocity (Vmax) and the Michaelis-Menten constant (Km). Vmax and Km from the Michaelis-Menten plot (Figure 3), were 0.10 µmol/min and 0.27 mM, respectively. We also plotted these data with several linear plots (data not shown) which confirmed the experimental Vmax and Km.
The values of the present study agree somewhat well with those of Surchandra et al., (2012)’s Vmax value of 1.33 µmol/min and better with their Km value of 0.416 mM. Other mung bean AP studies reported a Km of 0.3 mM (Nadir et al., 2012) and of 0.5 mM (Saeed et al., 2014), also which agree well with our Km value, especially when considering some studies were done with partially purified enzyme preparations. Additionally, a large difference was noticed in reaction rates when using acetate buffer versus citrate buffer. The enzyme reaction rate was about two times higher when run in 0.1 M acetate buffer, pH 5.2, compared to 0.1 M citrate, pH 5.2. Interestingly, Surchandra et al., (2012) routinely used citrate buffers in their assays. Along with other research groups, this study performs the AP assay in acetate buffer (Nadir et al., 2012; Saeed et al., 2014) because citrate has been known to inhibit acid phosphatase since 1969 (Verjee, 1969). Another possibility for differences in Vmax values is the number used for the molar absorptivity of the product, 4-nitrophenol (4NP). This study obtained 19.3 mM-1 cm-1 which was determined by measuring pure 4 NP in 0.1 M acetate buffer, pH 5.2. Surchandra et al. (2012) does not report what molar absorptivity was used, but in our hands the molar absorptivity of 4 NP in 0.1 M citrate buffer, pH 5.2, was about 47X lower than what it is in acetate. Finally, freshly sprouted mung beans were typically used and germinating seeds were not removed; we are not sure what part of the germinating seeds other investigators used.
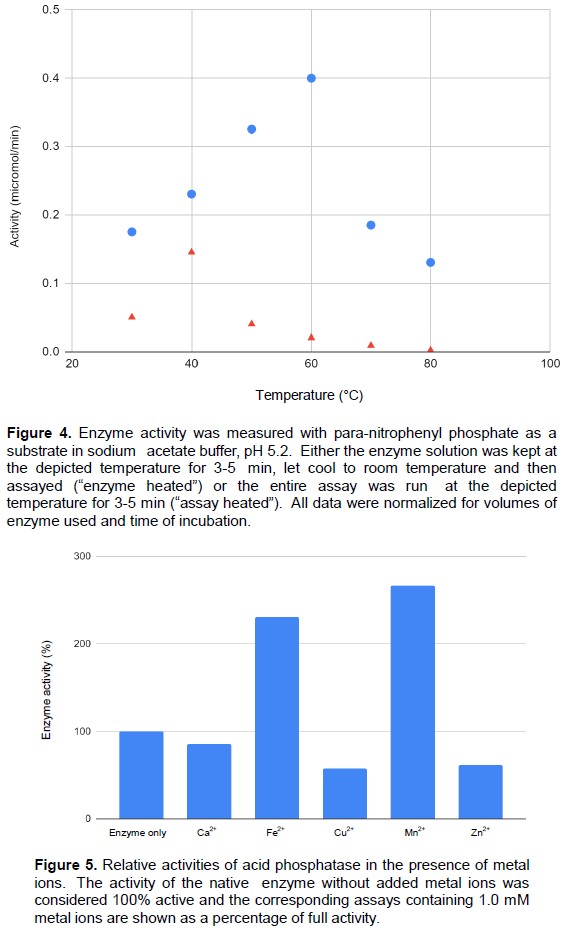
Heat sensitivity of acid phosphatase
It is interesting that mung bean AP is highly thermostable. Its activity was investigated at several temperatures and optimal temperature was found to be around 60°C (Figure 4), if we ran the entire assay at elevated temperatures. However, when the enzyme was heated first and then assayed there was a dramatic decrease in activity, even at the modest temperatures (Figure 4). At this point we do not have an explanation for the difference in activity. Our optimum temperature for activity of 60°C agrees well with most studies on mung bean acid phosphatase (Surchandra et al., 2012; Nadir et al., 2012; Saeed et al., 2014), but the exact optimal temperature for enzyme activity may not be a useful parameter (Almeida and Marana, 2019) and it might be better to classify enzymes as thermostable or non-thermostable.
Others have found phosphatase enzymes resistant to heat (Tagad and Sabharwal, 2018; Yenigün and Güvenilir, 2003; Wyss et al., 1998; Menichetti et al., 2015). We can speculate that naturally germinating seeds may experience extreme ranges of temperature and this heat stability trait is a survival advantage for plants.
Metal sensitivity
Some preliminary studies examining metal sensitivity show AP is activated by the presence of Mn2+ and Fe2+, while it is inhibited by Cu2+, Zn2+ and Ca2+ compared to buffer conditions (Figure 5). Metal dependence is known for certain acid phosphatases, although it is thought that the substrate is actually the moiety interacting with the metal ion rather than the enzyme itself (Brautigan, 2013).
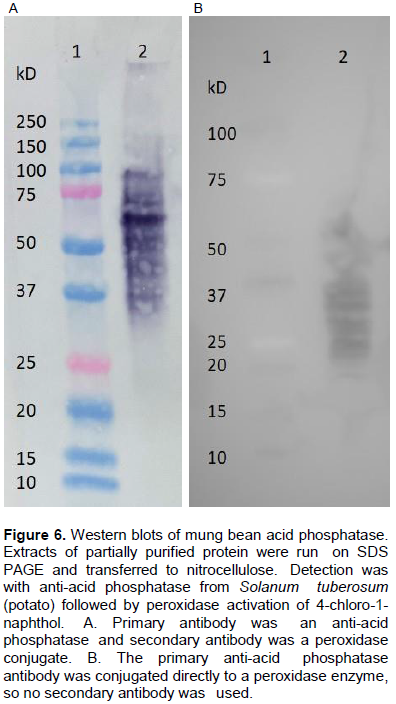
Western blot analysis
During this study, an antibody raised against mung bean AP was not available, but two different commercial anti-AP antibodies were used to begin investigating the structural similarities between plant acid phosphatases. Using a primary antibody against S. tuberosum (potato), the antibody was found to recognize several products from partially purified mung bean AP (Figure 6A, lane 2). The most recognized protein by the potato antibody is about 68 kDa and not our purified AP. Surprisingly, this antibody does not recognize any proteins in the molecular weight range of the mung bean AP. A different anti-potato AP antibody was used in a preliminary study (a peroxidase conjugate), and while it also recognizes many proteins, the detected proteins are in the appropriate molecular weight range (Figure 6B, lane 2). Future attempts will be made to use this antibody conjugate against the purified AP once we have obtained enough protein for it to be detectable.
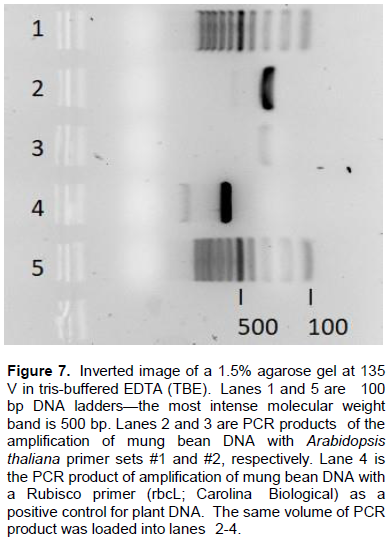
Genetic analysis suggests AP may function as a vegetative storage protein
DNA primers were designed based on the nucleotide sequence of A. thaliana vegetative storage protein 2 (VSP2) found in the NCBI protein/gene database, because at the time of this study, no sequences for any V. radiata genes had been deposited. Amplification with A. thaliana primers (set #1) led to one major band at just under 300 bp (Figure 7, lane 2). Another set of A. thaliana primers (set #2) led to one much weaker band at approximately 300 bp (Figure 7, lane 3). As a positive control, a Ribulose-1,5-bisphosphate carboxylase/oxygenase enzyme (Rubisco; rbcL) commercially-available primer was also used to amplify mung DNA. The PCR product is clearly visible at approximately 600 bp (Figure 7, Lane 4). The rbcL primer produces a 600 bp PCR product for Rubisco so these results verify high quality plant DNA from our mung bean extraction.
The only difference between the two primers sets used in lanes 2 and 3 (Figure 7) was an additional nucleotide A. Both primer sets were designed to hybridize to the flanking nucleotide regions of the three exons of A. thaliana VSP2, located on chromosome 5.
Because the mung bean DNA actually amplified with A. thaliana primers suggests that the VSP2 acid phosphatase gene sequence in A. thaliana is related to something in V. radiata. A PCR product of 300 bp suggests a protein of about 100 amino acids, while the A. thaliana VSP2 is 265 amino acids (Swarbreck et al., 2011). After our genetic results were obtained, a predicted acid phosphatase protein for mung bean was deposited in the NCBI database (accession #XP_014490817.1) (Kang et al., 2014). The translated amino acid sequence for mung bean AP contains the DXDXT consensus sequence (Collet et al., 1998) as well as an additional GDXXXD consensus sequence (Thaller et al., 1998) and has an expected mass of 30, 945 Daltons. This predicted protein fits our purified mung bean AP. Comparison of the mung bean predicted AP protein sequence with the A. thaliana VSP2 protein sequence shows only a 41% sequence identity, but the VSP2 protein also contains the same two AP consensus sequences of DXDXT and GDXXXD. Taken together these results suggest the mung bean AP and A. thaliana VSP2 are related, and that mung bean AP may have another role as a vegetative storage protein. It was a surprise that our mung bean PCR amplification did not lead to a larger product; it was well short of the 2000 bp expected due to the location of the primers (perimeter of the A. thaliana exons). The low sequence identity between the two proteins could explain this. It will be interesting to see what part of the mung bean exon/intron region amplified, and our next step will be to sequence our mung bean PCR product. A. thaliana VSP2 has also been found to exert anti-insect activity through its acid phosphatase consensus sequence of DXDXT (Liu et al., 2005). As the predicted protein for mung bean AP has this consensus sequence, it may have a third role as an anti-insect protein. Future studies will be needed to confirm these additional roles for mung bean AP.
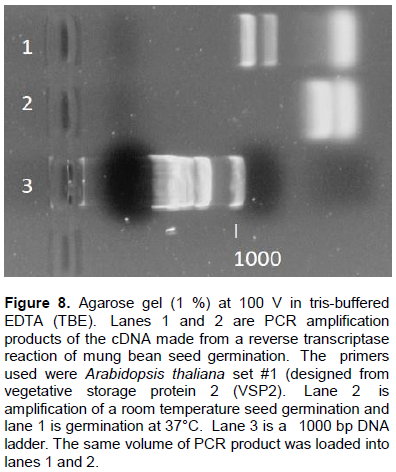
Effect of heat on AP protein synthesis
Since mung bean AP is heat stable we were interested if expression of proteins varied during seed germination if the temperature was raised. Seeds were germinated at room temperature and at 37°C and RNA transcription was compared. With the A. thaliana primer set #1 we used reverse transcription to create cDNA from isolated mRNA and compared these on a gel (Figure 8). Lane 1 shows cDNA generated from V. radiata seed germination at 37°C. There are two distinct bands, one at about 750 bp, and the lower one about 500 bp (based on multiple gels and molecular weight ladders), as well as a larger “fuzzy” band at less than 200 bp (Figure 8, Lane 1). Lane 2 of this gel picture contains the cDNA from a room temperature seed germination of V. radiata. Clearly, there are no higher molecular weight cDNA bands, only the lower than 200 bp “fuzzy” bands. The lower bands are indicative of primer aggregation, and do not suggest successful transcription. Because there is evidence of transcription in lane 1 (750 and 500 bp bands), it may be that mRNA transcription of a VSP2 homolog in mung bean during seed germination is induced at elevated temperatures. Thus, it was concluded that the mung bean VSP2 homolog might aid the plant during extreme temperatures.
An attempt was made to duplicate an ammonium sulfate precipitation purification of mung bean acid phosphatase reported previously (Surchandra et al., 2012) which led to a pure 34.5 kD enzyme. In this study, the published procedure led to a mixture of proteins which could not further be resolved by typical protein purification techniques. Mung bean acid phosphatase was purified using heat denaturation or band excision from an SDS polyacrylamide gel. Measuring Vmax and Km with 4-nitrophenylphosphate as the substrate on the partially purified mung bean extract yielded 0.10 µmol/min and 0.27 mM, respectively. These values are comparable to those obtained by others (Surchandra et al., 2012; Nadir et al., 2012; Saeed et al., 2014). Using primers designed against an A. thaliana AP-vegetative storage protein/anti-insect gene, a short V. radiata gene was amplified. This suggests there is at least one VSP2 homolog in mung bean, further studies will be required to determine if the mung bean protein similarly has insecticide activity. As the anti-potato antibody immunologically recognized several mung bean proteins, it is clear that plant acid phosphatases are structurally related, and additional research will address how much they are functionally-related.
The increase in mung bean mRNA during elevated temperature implies that AP thermal stability is not a coincidence and rather it is an important aspect of plant survival during potential temperature extremes. These results show that acid phosphatases are an interesting class of enzymes in plants and understanding them better may lead to advances not only in enzyme characterization, but in agricultural practices.
The authors have not declared any conflict of interests.
REFERENCES
|
Almeida VM, Marana SR (2019). Optimum temperature may be a misleading parameter in enzyme characterization and application. PLoS ONE 14(2):e0212977.
Crossref
|
|
|
|
Anand A, Srivastava PK (2012). A molecular description of acid phosphatase. Applied Biochemistry and Biotechnology 167(8):2174-2197.
Crossref
|
|
|
|
|
Anand A, Srivastava PK (2014). Isolation and enzymatic properties of a nonspecific acid phosphatase from Vigna aconitifolia seeds. Biotechnology and Applied Biochemistry 61(2):145-152.
Crossref
|
|
|
|
|
Asaduzzaman AK, Rahman HM, Yeasmin T (2011). Purification and characterization of acid phosphatase from a germinating black gram (Vigna mungo L.) seedling. Archives of Biological Sciences 63(3):747-756.
Crossref
|
|
|
|
|
Baldwin JC, Karthikeyan AS, Raghothama KG (2001). LEPS2, a phosphorus starvation-induced novel acid phosphatase from tomato. Plant Physiology 125(2):728-737.
Crossref
|
|
|
|
|
Bargaz A, Ghoulam C, Devron JJ (2013). Specific expression and activity of acid phosphatases in common bean nodules. Plant Signaling and Behavior 8(8):e25022.
Crossref
|
|
|
|
|
Bheri M, Mahiwal S, Sanyal SK, Pandey GK (2021). Plant protein phosphatases: What do we know about their mechanism of action? The FEBS Journal 288(3):756-785.
Crossref
|
|
|
|
|
Brautigan DL (2013). Protein Ser/Thr phosphatases - the ugly ducklings of cell signalling. The FEBS Journal 280(2):324-345.
Crossref
|
|
|
|
|
Collet JF, Stroobant V, Pirard M, Delpierre G, van Schaftingen E (1998). A new class of phosphotransferases phosphorylated on an aspartate residue in an amino-terminal DXDX(T/V) motif. Journal of Biological Chemistry 273(23):14107-14112.
Crossref
|
|
|
|
|
Copeland RA (2000). Enzymes, A practical introduction to structure, mechanism, and data analysis. 2nd edition, Wiley-VCH, New York, NY, pp. 40, 132, 134-135.
|
|
|
|
|
Davis L, Kuehl M, Battey J (1994). Basic methods in molecular biology. 2nd edition, Appleton & Lange, East Norwalk, CT, pp. 159-161, and 750.
|
|
|
|
|
Duff SMG, Sarath G, Plaxton WC (1994). The role of acid phosphatases in plant phosphorus metabolism. Physiologia Plantarum 90(4):781-800.
Crossref
|
|
|
|
|
Gonnety JT, Niamke S, Faulet BM, Kouadio EJPN, Kouame LP (2006). Purification and characterization of three low-molecular-weight acid phosphatases from peanut (Arachis hypogaea) seedlings. African Journal of Biotechnology 5(1):035-044.
|
|
|
|
|
Hegeman OCE, Grabau EA (2001). A novel phytase with sequence similarity to purple acid phosphatases is expressed in cotyledons of germinating soybean seedlings. Plant Physiology 126(4):1598-1608.
Crossref
|
|
|
|
|
Joyce BK, Grisolia S (1960). Purification and properties of a nonspecific acid phosphatase from wheat germ. Journal of Biological Chemistry 235(8):2278-2281. PMID: 14408027.
Crossref
|
|
|
|
|
Kang JY, Kim SK, Kim M, MY, Lestari P, Kim KH, Ha BK, Jun TH, Hwang WJ, Lee T, Lee J, Shim S (2014). Genome sequence of mung bean and insights into evolution within the Vigna species. Nature Communications 5(5443).
Crossref
|
|
|
|
|
Laemmli UK (1970). Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227(5259):680-685.
Crossref
|
|
|
|
|
Li D, Zhu H, Liu K, Liu X, Leggewie G, Udvardi M, Wang D (2002). Purple acid phosphatases of Arabidopsis thaliana. Comparative analysis and differential regulation by phosphate deprivation. Journal of Biological Chemistry 277(31):27772-27781.
Crossref
|
|
|
|
|
Liu Y, Ahn JE, Datta S, Salzman RA, Moon J, Huyghues-Despointes B, Pittendrigh B, Murdock LL, Koiwa H, Zhu-Salzman K (2005). Arabidopsis vegetative storage protein is an anti-insect acid phosphatase. Plant Physiology 139(3):1545-1556.
Crossref
|
|
|
|
|
Menichetti L, Reyes Ortigoza A, García N, Giagnoni L, Nannipieri P, Renella G (2015). Thermal sensitivity of enzyme activity in tropical soils assessed by the Q10 and equilibrium model. Biology and Fertility of Soils 51:299-310.
Crossref
|
|
|
|
|
Mission J, Raghothama KG, Jain A, Jouhet J, Block MA, Bligny R, Ortet P, Creff A, Somerville S, Rolland N, Doumas P, Nacry P, Herrerra-Estrella L, Nussaume L, Thibaud MC (2005). A genome-wide transcriptional analysis using Arabidopsis thaliana Affymetrix gene chips determined plant responses to phosphate deprivation. Proceedings of the National Academy of Sciences 102(33):11934-11939.
Crossref
|
|
|
|
|
Moorman V, Brayton A (2021). Identification of individual components of a commercial wheat germ acid phosphatase preparation. PLoS ONE 16(3):e0248717.
Crossref
|
|
|
|
|
Nadir S, Saeed A, Naz R, Siddiqua A, Sherazi M, Wazir SM, Saeed A (2012). Isolation, purification and characterization of acid phosphatase from germinating Vigna radiata seeds. Journal of the Chemical Society of Pakistan 34(3):717-727.
|
|
|
|
|
Olczak M, Watorek W, Morawiecka B (1997). Purification and isolation of acid phosphatase from yellow lupin seeds. Biochimica et Biophysica Acta 1341(1):14-25.
Crossref
|
|
|
|
|
Saeed A, Salim M, Naz R, Zaman U, Baloch AL, Nadir S, Saeed A (2014) Partial purification, characterization and some kinetic properties of low molecular weight acid phosphatase from leaves of germinating Vigna radiata seeds. The Journal of Animal and Plant Sciences 24(5):1466-1477.
|
|
|
|
|
Schenk G, Guddat LW, Ge Y, Carrington LE, Hume DA, Hamilton S, de Jersey J (2000). Identification of mammalian-like purple acid phosphatases in a wide range of plants. Gene 250(1-2):117-125.
Crossref
|
|
|
|
|
Schweighofer A, Meskiene I (2015). Phosphatases in plants. Methods in Molecular Biology 1306:25-46.
Crossref
|
|
|
|
|
Staswick PE (1994). Storage proteins of vegetative plant tissues. Annual Review of Plant Physiology and Plant Molecular Biology 45(1):303-322.
Crossref
|
|
|
|
|
Surchandra T, Roy SS, Singh NR, Sahoo MR, Prakash N (2012). Partial purification and biochemical characterization of acid phosphatase from germinated mung bean (Vigna radiata) seeds. African Journal of Biotechnology 11(103):16777-16782.
|
|
|
|
|
Swarbreck D, Lamesch P, Wilks C, Huala E (2011). Direct submission to Arabidopsis TAIR10 release. The Arabidopsis Information Resource, Department of Plant Biology, Carnegie Institution, Stanford, CA, 2011.
|
|
|
|
|
Tagad CK, Sabharwal SG (2018). Purification and characterization of acid phosphatase from Macrotyloma uiflorum seeds. Journal of Food Science Technology 55(1):313-320.
Crossref
|
|
|
|
|
Tejera Garcia NA, Olivera M, Iribarne C, and Lluch C. (2004) Partial purification and characterization of a non-specific acid phosphatase in leaves and root nodules of Phaseolus vulgaris. Plant Physiology and Biochemistry 42(7-8):585-591.
Crossref
|
|
|
|
|
Thaller MC, Schippa S, Rossolini GM (1998). Conserved sequence motifs among bacterial, eukaryotic, and archaeal phosphatases that define a new phosphohydrolase superfamily. Protein Science 7(7):1647.
Crossref
|
|
|
|
|
The Arabidopsis Genome Initiative (2000). Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408:796-815.
Crossref
|
|
|
|
|
Tran HT, Hurley BA, Plaxton WC (2010). Feeding hungry plants: The role of purple acid phosphatases in phosphate nutrition. Plant Science 179(1-2):14-27.
Crossref
|
|
|
|
|
Uhrig GR, Labandera AM, Moorhead GB (2013). Arabidopsis PPP family of serine/threonine protein phosphatases: many targets but few engines. Trends in Plant Science 18(9):505-513.
Crossref
|
|
|
|
|
Verjee ZHM (1969). Isolation of three acid phosphatases from wheat germ. European Journal of Biochemistry 9(3):439-444.
Crossref
|
|
|
|
|
Waymack PP, Van Etten RL (1991). Isolation and characterization of a homogeneous isoenzyme of wheat germ acid phosphatase. Archives of Biochemistry and Biophysics 288(2):621-633.
Crossref
|
|
|
|
|
Wu P, Ma L, Hou X, Wang M, Wu Y, Liu F, Deng XW (2003). Phosphate starvation triggers distinct alterations of genome expression in Arabidopsis roots and leaves. Plant Physiology 132(3):1260-1271.
Crossref
|
|
|
|
|
Wyss M, Pasamontes L, Rémy R, Kohler J, Kusznir E, Gadient M, Müller F, van Loon AP (1998). Comparison of the thermostability properties of three acid phosphatases from molds: Aspergillus fumigatus phytase, A. niger phytase, and A. niger pH 2.5 acid phosphatase. Applied Environmental Microbiology 64(11):4446-4451.
Crossref
|
|
|
|
|
Xie L, Shang Q (2018). Genome-wide analysis of purple acid phosphatase structure and expression in ten vegetable species. BMC Genomics 19:646.
Crossref
|
|
|
|
|
Yenigün B, Güvenilir Y (2003). Partial purification and kinetic characterization of acid phosphatase from garlic seedling. Applied Biochemistry and Biotechnology 105-108:677-87.
Crossref
|
|
|
|
|
Zaman U, Naz R, Khattak NS, ur Rehman K, Iqbal A, Ahmad S, Shah LA (2020). Investigating the thermodynamic and kinetics properties of acid phosphatase extracted and purified from seedlings of Chenopodium murale. International Journal of Biological Macromolecules 165:1475-1481.
Crossref
|
|