ABSTRACT
Tinospora cordifolia is a highly important medicinal plant in India, with a rich source of different secondary metabolites. In the present study, 12 samples of T. cordifolia (collected from different areas of Rayalseema region,Andhra Pradesh) were studied for genetic diversity using 15 random amplified polymorphic DNA (RAPD) and eight inter-simple sequence repeats (ISSR) markers. Out of them, 10 RAPD and 5 ISSR primers showed 122 polymorphic reproducible bands across the selected 12 samples of T. cordifolia. The average polymorphic index content (PIC) of RAPD and ISSR values was 0.27 to 0.41 and 0.17 to 0.41, respectively. The PIC with RAPD markers was the highest for primers OPV 17 (0.41) and OPW 17(0.41) followed by the primer OPB 20 (0.40). The PIC with ISSR markers was the highest for the primer 824 (0.41) followed by primer 867 (0.36). Unweighted pair group method with arithmetic mean (UPGMA) cluster analysis of genetic similarity indices grouped all the samples into two major clusters. Jacquard’s coefficient of similarity varied from 0.51 to 0.77, indicating high levels of genetic variation across genotypes under study. The result of this study can be used for characterization of potential T. cordifolia genetic resources and their utilization as breeding materials, micropropagation and secondary metabolites screening and harvesting. T. cordifolia, being a medicinal plant, is very useful for discovery of new lead molecules from this plant for drug discovery and development studies.
Key words: Tinospora cordifolia, genetic diversity, random amplified polymorphic DNA (RAPD), inter-simple sequence repeats (ISSR), unweighted pair group method with arithmetic mean (UPGMA).
Tinospora cordifolia (Willd.), belonging to the family Menispermaceae is an important climber medicinal plant species found at an altitude of 300 m, extending from the Himalayas down to the southern part of Peninsular India. The family Menispermaceae comprises about 72 genus and 450 species found in the tropical low land. Tinospora is one of the important genus of this family, consisting of about 32 species distributed in tropical Africa, Madagascar, Asia to Australia and the Pacific Islands (Kubitzki et al., 1993; Mabberley, 1997). It is also found in China, Myanmar, Sri Lanka, Thailand, Philippines, Indonesia, Malaysia, Borneo, Vietnam, Bangladesh, Pakistan, North Africa, West Africa and South Africa (Singh et al., 2003; Mia et al., 2009; Jain et al., 2010). The wood is white, soft, and porous. Leaves are simple, alternate, exstipulate, long petiolate, chordate shaped, long thread-like aerial roots arising from the branches with small unisexual yellow or greenish yellow flowers (Anonymous; Wealth of India, 1976; Kirthikar and Basu 1975). Fruits are red, fleshy, with many drupelets on a thick stalk. Three major groups of compounds; protoberberine alkaloids, terpenoids and polysaccharides are considered as putative active constituents in T. cordifolia (Chintalwar et al., 1999; Bisset and Nwaiwu, 1983). The stem is bitter, stomachic, diuretic (Nayampalli et al., 1988), stimulates bile secretion, causes constipation, allays thirst, burning sensation, vomiting, enriches the blood and cures jaundice, T. cordifolia is extensively used in the Indian Ayurvedic system of medicine as a tonic and also exhibits anti-periodic, antispasmodic, anti-inflammatory anti-arthritic, anti-allergic activities and also is very popular for anti-diabetic treatment (Handique, 2014) and also used in many ayurvedic formulations for treatment of different infections (Shanti and Nelson, 2013).T. cordifolia shows good immunomodulatory response (Jitendra et al., 2014). The stem of T. cordifolia is used as medicine as reported in Ayurvedic Pharmacopoeia of India (Sivakumar et al., 2014). The extract of its stem is useful in skin diseases (Aiyer and Kolammal, 1963; Raghunathan and Mittra, 1982). The root and stem of T. cordifolia are prescribed in combination with other drugs as an antidote to snake bite and scorpion sting (Nadkarni and Nadkarni, 1976; Kirtikar and Basu, 1975; Zhao et al., 1991). The plant is very rigid and grows in almost all climates especially preferring warm climate. Planting is usually done during the rainy season (July to August).
The DNA profiling technique using Random amplified polymorphic DNA (RAPD) and inter-simple sequence repeats (ISSR), is extensively used in plants for the study of biodiversity, gene identification, and identifying markers linked with traits of interest. Plant genetic stability study using molecular markers in medicinal plants like Withania somnifera and Capparis deciduas (Tyagi et al., 2010) and other plants like coffee species (Mishra et al., 2014), artichoke (Sharaf-Eldin et al., 2015), bamboo (Desai et al., 2015), rye grass (Ghariani et al., 2015), and lemongrass (Bishoyi et al., 2016) has been reported.
Irrespective of the plant source or age, RAPD patterns are consistent and very useful for germplasm characterization, estimation of genetic relatedness and conservation of plant genetic resources (Welsh and McClelland, 1990). ISSR markers are also PCR based. It involves amplification of DNA segments between two identical microsatellite repeat regions with high reproducibility due to the use of longer primers (16 to 25-mer), and its high annealing temperature (45 to 60°C) leads to higher stringency (Zietkiewicz et al., 1994; Pradeep Reddy et al., 2002). ISSR has been found to be a powerful, rapid, simple, economical and reproducible tool used in marker assisted selection, DNA fingerprinting, evolution and molecular ecology (Zhao et al., 2007; Gajera et al., 2010; Zhang and Dai, 2010). Based on the advantages of these two molecular markers, viz., RAPD and ISSRs, they were employed to assess the genetic variation at the molecular level in T. cordifolia (Willd.) collected from the Rayalaseema region of Andhra Pradesh.
T. cordifolia has been listed amongst 29 highly prioritized medicinal plants for agro climatic zone 8 (Rajasthan, Uttar Pradesh and Madhya Pradesh) of India as identified by the National Medicinal Plant Board (NMPB) New Delhi, Government of India. This plant has also been listed in 178 medicinal plant species in high volume trade by NMPB, New Delhi, India (National Medicinal Plant Board, 2014). Due to the enormous traditional, folk, ayurvedic, medicinal and phytochemical importance, this plant species are disappearing rapidly from their natural habitats and is becoming endangered. There are no reports on studies of molecular profiling with molecular markers reported from the Rayalseema region of Andhra Pradesh. Assessment of genetic diversity will help in identifying elite trees with superior traits and also help in correlation of genetic diversity with function. This will be useful for conserving morphological and biological diversity of a population.
Plant collection
A total of 12 samples of T. cordifolia were collected from different places of the YSR Kadapa district and Kurnool districts of Rayalaseema region, Andhra Pradesh having an altitude ranging from 120 m to around 500 m, and latitude ranging from 14 to 15, and longitudes at 77 to 79 range as listed in Table 1 and they were used in the present study. The plants were authenticated in Department of Botany, Yogi Vemana University.
DNA extraction
Total genomic DNA was extracted from young leaves using modified CTAB method (Murray and Thompson, 1980). Approximately, 200 mg of young leaf tissue devoid of mid rib was taken and ground to fine powder in liquid nitrogen in a motor and pestle. The finely ground powder was then transferred to 2.0 ml Eppendorff and to this 1 ml of preheated 2X CTAB buffer containing 2% (w/v) CTAB, 1.4 M NaCl, 20 Mm EDTA, 100 mM Tris-HCl, pH 8.0 was added. To this mixture, 0.2% (v/v) -mercaptoethanol was added and the homogenate was incubated at 65°C for 90 min with constant gentle mixing. Later, the homogenate was cooled to room temperature and equal volume of chloroform:isoamyl alcohol (24:1) was added and centrifuged at 12,000 rpm for 10 min at 25°C. Supernatant was collected and to this 10 µl of RNase (10 mg/ml) was added and incubated at 37°C for 30 min. To this, equal volumes of chloroform:isoamylalcohol (24:1) were added and extracted to remove any remains of proteins by centrifuging at 12,000 rpm for 10 min at 25°C (Eppendroff Centrifuge 5810-R). The supernatant was then transferred to fresh 1.5 ml Eppendorff, and to which 0.6 ml of ice cold isopropanol was added and incubated at -20°C for 30 min. The DNA is pelleted out by centrifuging at 12,000 rpm for 10 min, washed with 70% ethanol, air dried and dissolved in sterile double distilled water. Quantification of the genomic DNA was done by spectrophotometric measurement of UV absorbance at 260 nm (UV 1800 ShimadzuUV spectrophotometer). An aliquot of the DNA samples was diluted in sterile distilled water in a ratio of 1:1000 in a 1-ml cuvette and the optical density was determined at 260 and 280 against sterile distilled water as blank. The DNA concentration was calculated using the formula (Sambrook et al., 1989):
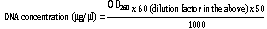
The ratio of OD260 to OD280 was calculated to check the purity of DNA. DNA samples for analysis were diluted to 7.5 ng/µl for RAPD and ISSR analysis.
Analysis with RAPD markers
A set of 15 RAPD (decamer) primers was randomly selected and used for PCR amplification (Table 2). PCR reaction was performed in a volume of 20 µl reaction mix containing 10 mM Tris-HCl, 1.5 mM MgCl2, 5 pM of decamer primer, 250 µM each dNTP’s (Biobasic, Canada) and 0.8 U of TaqDNA polymerase with 30 ng of genomic DNA. PCR amplification was performed on thermocycler (Eppendorf Master Cycler Pro AG 6321, Germany). With initial denaturation at 95°C for 3 min, followed by 40 cycles of denaturation at 94°C for 1 min, 37°C annealing temperature for 1 min, extension time of 1 min at 72°C, followed by final extension at 72°C for 10 min. The amplicons were resolved on 1.2% agarose gel in 0.5x TBE system with a constant voltage of 100V for 2 h. A 1 kb DNA ladder (Hi-Media, India) was included as a control size marker. The amplified DNA was stained with Ethidium bromide, observed and documented in Gel documentation unit (G Box Syngene, Synoptic, Ltd, U.K).
Analysis with ISSR markers
A total of eight ISSR primers were employed for PCR amplification (Table 3). PCR reaction was performed in a volume of 20 µl reaction mix containing 10 mM Tris-HCl, 1.5 mM MgCl2, 5 pM of decamer primer, 250 µM each dNTP’s (Biobasic, Canada) and 0.8 U of Taq DNA polymerase with 30 ng of genomic DNA. PCR amplification was performed on thermocycler (Eppendorf Master Cycler Pro AG 6321, Germany). With initial denaturation at 95°C for 3 min, followed by 40 cycles of denaturation at 94°C for 1 min, annealing at 45°C for 1 min, extension time of 1 min at 72°C following final extension at 72°C for 10 min. The amplicons were resolved on 1.2% agarose gel in 0.5x TBE system with a constant voltage of 100 V for 2 h. A 1 kb DNA ladder (Hi-Media Labs, India) was included as a control size marker. The DNA was stained with Ethidium bromide, observed and documented in Gel documentation unit (G Box Syngene, Synoptic, Ltd, U.K).
Data scoring and analysis
The reaction was carried out twice and the gel was checked for consistency of the polymorphic bands. The reproducible bands were scored in all the samples for 15 RAPD and 8 ISSR primers separately. Each amplified product was considered as a separate marker. The band profiles were scored only for distinct, reproducible bands after PCR for each of the RAPD and ISSR primers. The reproducible amplicons were scored as 1 for the presence of a band and 0 for its absence, respectively. Similarity matching (SM) DICE coefficient values for pair-wise comparison between accessions were calculated and a DICE coefficient matrix was constructed using the SIMQUAL subroutine. Jaccard’s similarity coefficient values were calculated and dendrograms based on similarity coefficient values were generated using unweighted pair-group method with arithmetic means (UPGMA) by the NTSYSpc 2.02j software. The polymorphism information content (PIC) value was calculated using the formula: 2f(1-f), where f is the frequency of bands present and (1- f), is the frequency of bands absent.
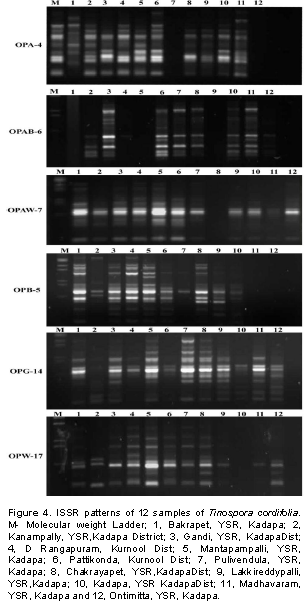
Screening of 12 T. cordifolia samples from the Rayalseema region with 15 RAPD and 8 ISSR markers, showed amplified amplicons with 10 RAPD primers and 5 ISSR primers. RAPD and ISSR markers showed 55 and 54% of polymorphism, respectively. These decamer RAPD primers OPB-20 and OPB-5 gave minimum of 4 amplicons and maximum of 12 amplicons, respectively each ranging from 300 to 1100 bp. From the total of 8 ISSR primers tested, five showed amplification. These five ISSR primers were able to amplify a minimum of four to eleven amplicons across the accessions with an average PIC value of 0.17 (846) to 0.41 (824) among the accessions (Table 4). The PCR amplified products of RAPD and ISSR primers are shown in Tables 2 and 3. The amplified product ranged between 0.35 and 1.8 kb in size for RAPD’s markers (Figure 3), while with ISSR primers the amplified product size ranged between 0.5 and 2 kb (Figure 4), respectively. In this study, the number of alleles generated for individual loci varied from four to twelve, hence explaining the PIC range between 0.27 and 0.41 with RAPD markers and 0.17 to 0.41 with ISSR markers, respectively.
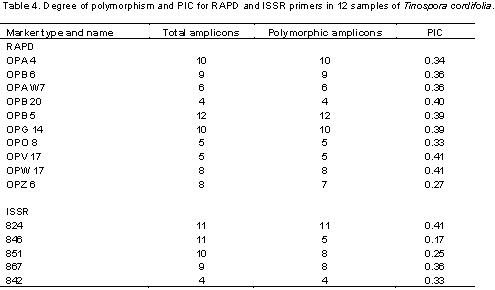
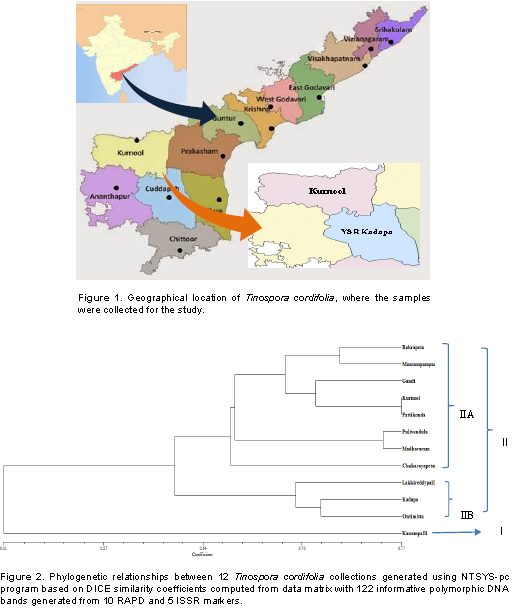
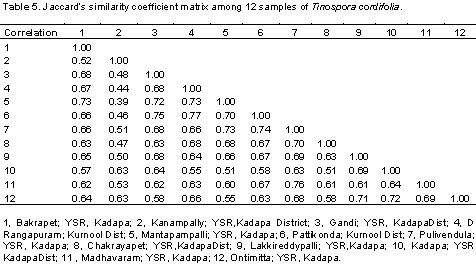
A total of 122 polymorphic bands were scored from 10 RAPD and 5 ISSR primers. A dendrogram was constructed to reveal the genetic relationships among the T. cordifolia (Figure 1). The UPGMA-based dendrogram showed that, all of the 12 collections could be classified into two groups based on the 122 polymorphic amplicons between the samples. The genetic distances among 12 genotypes are shown in Table 4. The similarity values in terms of genetic distance ranged from 0.51 to 0.77. A dendrogram generated by cluster analysis (UPGMA method) based on the 122 polymorphic amplicons revealed that the total 12 collections were separable into two major clusters, Cluster-I and Cluster-II (Figure 2). Cluster-I includes only one collection made from the region of Kanampalli near Pulivendula of YSR-Kadapa district, while the rest 11 collections are grouped into Cluster-II. These 11 collections from the Cluster-II were grouped into two groups that include eight collections into Cluster-IIA while three collections in Cluster-IIB. The same has been reflected in the similarity table, where the similarity coefficient of the sample from Kanampalli is distinct from others with less similarity range of 0.39 to 0.63 compared to collection from Pattikonda and Kurnool with a high similarity range of 0.63 to 0.77 in comparison with other genotypes under study.
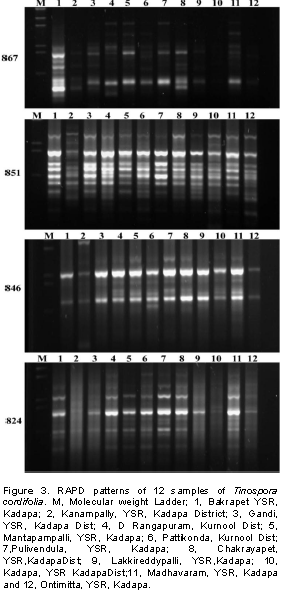
Though RAPD markers have the problem of reproducibility, until recently and till to date these are the proven markers in terms of cost effectiveness that were extensively used to study genetic diversity among the species of bacteria, fungi, insects, plants and animals. Genetic diversity of twelve populations of three mushroom species were evaluated using seven RAPD markers that were able to differentiate the percent variability within and among different populations of these mushroom species (Dwivedi et al., 2018). Sugarcane when genetic diversity was estimated contain similarity coefficient which ranged from 0.43 to 0.91 using thirty five RAPD markers while the ISSR primers could explain the similarity coefficient from 0.73 to 0.93 which is less compared to RAPD markers (Patel et al., 2018). Eighteen synthetic Elite-II hexaploid wheat lines were evaluated using RAPD and SSR based molecular markers and inferred genetic variability of these synthetic lines is due to the novel genetic background derived from Aegilops tauschi accessions used as parental lines (Ahmad, 2014). While twenty two RAPD markers were used to study genetic diversity and percent polymorphism of 20 cotton germplasm lines that can help in identification of novel parental lines for hybrid cotton development (Tidke et al., 2014). A total of 16 RAPD and six SSR markers were used to assess the genetic diversity within the Jatropha curcas lines of diverse origin along with two of its wild relatives and showed the effectiveness of these markers to differentiate the wild relatives from the cultivated lines of Jatropa (Reddi et al., 2016). Wang et al., (2016) effectively utilized 100 RAPD primers and evaluated population structure of ginseng (Panax ginseng C.A. Meyer) collected from different regions and estimated the extent of gene flow between populations while Chandrakala et al., (2017) had shown that combine application of RAPD and ISSR markers can be effectively utilize to study the molecular genetic diversity in Canthium parviflorum Lam a medicinally important plant with wide range of applications. Among RAPD, molecular markers have been successfully used in studying the degree of genetic diversity between the Morinda citrifolia (noni) plants on the Asian continent. Singh et al. (2012) and Patel et al. (2014) have revealed high genetic diversity in M. citrifolia plants in different regions of India. This was probably due to the fact that region was being considered the center of origin of this species.
In this study, the results showed that the sub clusters formed by plants of different location in Rayalseema region showed the level and extent of genetic variation present in them. The genetic distance similarity matrix among the 12 T. cordifolia collections is shown in Table 5. The similarity values in terms of genetic distance ranged from 0.39 to 0.77 between the samples from different places. Based on the molecular genetic studies and similarity index, samples from Kanampalli compared to any others were distant with an average similarity of 0.39 to 0.63 from other collections. Whereas collection from Kanampalli and Mantapampalli are more distant from each other with minimum similarity of only 0.39, collections from Kurnool and Pattikonda are much similar to each other with a similarity index of 0.77. It is also evident that from the dendrogram generated based on 122 number of amplicons, samples from Kanampalli of YSR-Kadapadistrict, Andhra Pradesh, India has turned out to be more distinct from the other 11 collections. It is justified by the fact that the geographical location of this place is much narrowly connected with the other places in the adjacent region, with a fertile valley surrounded by mountains, the population growing in this place is more distinct with less genetic similarity index compared with other collections.
The polymorphism data generated can be used for plant breeding, crop improvement programs and also might be helpful in future strategies for evaluation of desired genotypes. More investigation on Tinospora genetic diversity is needed to evaluate more intra specific diversity analysis. Such studies on intra specific genetic diversity can contribute to the development of conservation strategies by identifying units for conservation. John De Britto et al., (2010) studied and determined the genetic variability of T. cordifolia among the selected populations using RAPD markers and concluded the existence of low level of genetic variability in the species in a small geographical area among the different accessions of T. cordifolia. The number of polymorphic loci is 19 and the percentage of polymorphic loci is 46.34. The overall genetic distance and the genetic identity ranges from 0.1872 to 0.4555 and 0.6341 to 0.8293, respectively. The dendrogram showed three clusters between five populations and the studies revealed that wide heritable phenotypic and chemotypic variation in the collection of accessions might be due to qualitative genetic differences. Rana et al., (2012) demonstrated the genetic diversity analysis of T. cordifolia germplasm collected from northwestern Himalayan region of India.
The RAPD markers have also been employed in estimating genetic variability of various plants species. Molecular analysis in Urginea indica collected from different location of Karnataka was reported by Harini et al., (2008). Genetic diversity analysis in Rauvolfia serpentine and Rauvolfia tetra phyla L., using RAPD markers was carried out by Padmalatha and Prasad (2006, 2007). Ruan et al. (2008) analyzed the DNA molecular characters of Centella asiatica with RAPD techniques. The RAPD technique has been widely used both for studies on wild plants (Yeh et al., 1995; Khasa and Dancik, 1996; Manica-Cattani et al., 2009) and for studies on cultivated plants (Sharma and Dowsons, 1995; Yilmaz et al., 2012). In the present study, ISSR has been used as an effective molecular tool to calculate the genetic diversity on wild medicinal plant like T. cordifolia. Ahmed et al., (2006) utilized two molecular approaches, viz., 38 RAPD primers assay and restriction digestion of ITS1-5.8S-ITS2 Rdna (PCR-RFLP) in order to authenticate the results obtained by RAPD and PCR-RFLP to evaluate the genetic similarities between 40 different accessions belonging to three species (T. cordifolia, Tinospora malabarica and Tinospora crispa).
Three independent clones of each species were sequenced and phylogenetic relationship obtained from ITS sequences was found to be in agreement with RAPD data and inter and intra specific variation in the ITS region of Tinospora species that has been observed. The average proportion of polymorphic markers across primers was 95%, whereas restriction endonucleases showed 92% polymorphism. RAPD alone was found suitable for the species diversions. In contrast, PCR-RFLP showed bias in detecting exact species variation. There is limited information available for genus Tinospora where molecular markers like RAPD and ISSR were utilized for molecular genetic studies. The present analysis is based on ten RAPD and five ISSR markers which revealed significant genetic diversity among the T. cordifolia samples studied.
In this preliminary study of genetic diversity analysis which has not been reported earlier from this region, the existence of genetic diversity among a selected sample of T. cordifolia collected from various localities in Rayalseema region was reported. The results with RAPD showed that there is a need to take samples from wider populations situated at distant geographical locations to better assess their phylogenetic relationship so as to preserve their diversity for the future work. Identification of intra-population diversity also forms a very essential pre-requisite for the genetic diversity analysis. This is the first report of genetic diversity analysis in T. cordifolia from the Rayalseema region in Andhra Pradesh.
The authors have not declared any conflict of interests.
The first author who is an INSPIRE FELLOW (110388) is thankful to the Department of Science and Technology, Ministry of Science and Technology, Government of India, New Delhi for providing fellowship during the research work and Agri-Science Park for extending the infrastructural support.
REFERENCES
|
Agro-techniques of Selected Medicinal Plants,Vol.II, National Medicinal Plants Board, Department of AYUSH, Ministry of Health and Family Welfare, Government of India, 2014. ISBN No – 978-81-909121-3-6.
|
|
|
|
Ahmed SM, Verma V, Qazi PH, Ganaie MM, Bakshi SK, Qazi GN (2006). Molecular phylogeny in Indian Tinospora species by DNA based molecular markers. Plant Systematics and Evolution 256(1-4):75-87.
Crossref
|
|
|
|
|
Ahmad K (2014). RAPD and SSR based genetic diversity analysis of elite-2 set of synthetic hexaploid wheats. African Journal of Traditional, Complementary and Alternative Medicines 11(4):9-13.
Crossref
|
|
|
|
|
Aiyer KN, Kolammal M (1963). Pharmacognosy of Ayurvedic Drugs, Series 1. The Central Research Institute, Trivandrum.
|
|
|
|
|
Anonymous (1976). The Wealth of India. A dictionary of Indian raw materials. Vol. X., C.S.I.R.f New Delhi, India.
View
|
|
|
|
|
Bishoyi AK, Sharma A, Kavane A, Geetha KA (2016). Varietal discrimination and genetic variability analysis of Cymbopogon using RAPD and ISSR markers analysis. Applied Biochemistry and Biotechnology 179(4):659-670.
Crossref
|
|
|
|
|
Bisset N, Nwaiwu (1983). Quaternary alkaloids of tinospora species. Planta Medica 48(8):275-279.
Crossref
|
|
|
|
|
Chintalwar G, Jain A, Sipahimalani A, Banerji A, Sumariwalla P, Ramakrishnan R, Sainis K (1999). An immunologically active arabinogalactan from Tinosporacordifolia. Phytochemistry 52(6):1089-1093
Crossref
|
|
|
|
|
Chandrakala S, Reddi KVNR, Sekhar AC, Reddy PCO, Mallikarjuna K (2017). In vitro propagation and genetic diversity analysis of Canthium parviflorum Lam by RAPD and ISSR markers. Annals of Plant Sciences 6(11):1775-1783.
Crossref
|
|
|
|
|
Desai P, Gajera B, Mankad M, Shah S, Patel A, Patil G, Narayanan S, Kumar N (2015). Comparative assessment of genetic diversity among Indian bamboo genotypes using RAPD and ISSR markers. Molecular Biology Reports 42(8):1265-1273.
Crossref
|
|
|
|
|
Dwivedi S, Singh S, Chauhan, UK, Tiwari MK (2018). Inter and intraspecific genetic diversity (RAPD) among three most frequent species of macrofungi (Ganoderma lucidum, Leucoagricus sp. And Lentinus sp.) of Tropical forest of Central India. Journal of Genetic Engineering and Biotechnology 16(1):133-141.
Crossref
|
|
|
|
|
Ghariani S, Elazreg H, Chtourou-Ghorbel N, Chakroun M, Trifi-Farah N (2015). Genetic diversity analysis in Tunisian perennial ryegrass germplasm as estimated by RAPD, ISSR, and morpho-agronomical markers. Genetics and Molecular Research 14(4):18523-18533.
Crossref
|
|
|
|
|
Gajera BB, Kumar N, Singh AS, Punvar BS, Ravi kiran R, Subhash N, Jadeja GC (2010). Assessment of genetic diversity in castor (Ricinus communis L.) using RAPD and ISSR markers. Industrial Crops and Products 32(3):491-498.
Crossref
|
|
|
|
|
Handique PJ (2014). In Vitro Propagation and Medicinal Attributes of Tinospora Cordifolia: A Review. Austin Journal of Biotechnology and Bioengineering 1(5):5.
|
|
|
|
|
Harini SS, Leelambika L, Kameshwari S, Sathyanarayana N (2008). Optimization of DNA isolation and PCR-RAPD methods for molecular analysis of Urginea indica Kunth. International Journal of Integrative Biology 2(2):138-44.
|
|
|
|
|
Jain S, Sherlekar B, Barik R (2010). Evaluation of antioxidant potential of Tinospora cordifolia and Tinospora sinensis. International Journal of Pharmaceutical Sciences and Research 1(11):122-128.
|
|
|
|
|
Khasa PD, Dancik BP (1996). Rapid identification of white-Engelmann spruce species by RAPD markers. Theoretical and Applied Genetics 92(1):46-52.
Crossref
|
|
|
|
|
Jitendra Mittal, Madan Mohan Sharma, Amla Batra (2014). Tinospora cordifolia: a multipurpose medicinal plant- A review. Journal of Medicinal Plants Studies 2 (2):32-47.
|
|
|
|
|
John De Britto A, Leon Stephan Raj T, Petchimuthu K, Benjamin JeyaRathna, Kumar P, Mary Sujin R, Dharmar K (2010). Inter Population Genetic Variability in Tinosporacordifolia(wild.) Miers ex Hook. F. &Thoms .(Menispermaceae), through RAPD marker, Sciencia Acta Xaveriana 1(1):57-64.
|
|
|
|
|
Kirthikar KR, Basu BD (1975). Indian Medicinal Plants Vol-4. Bishen Singh Mahendra Pal Singh; Dehradun.
View
|
|
|
|
|
Kubitzki K, Rohwer JG, Bittrich V (1993). The families and genera of vascular plants. Vol.2. Flowering plants, dicotyledons: Magnoliid, Hamamelid, and caryophyllid families. https://doi.org/10.1007/978-3-662-07255-4
Crossref
|
|
|
|
|
Mabberley DJ (1997). The plant-book: a portable dictionary of the vascular plants. Cambridge University Press.
|
|
|
|
|
Manica-Cattani MF, Zacaria J, Pauletti G, Atti-Serafini L, Echeverrigaray S (2009). Genetic variation among South Brazilian accessions of Lippia alba Mill.(Verbenaceae) detected by ISSR and RAPD markers. Brazilian Journal of Biology 69(2):375-380.
Crossref
|
|
|
|
|
Mia MM, Kadir MF, Hossan MS. Rahmatullah M (2009). Medicinal plants of the Garo tribe inhabiting the Madhupur forest region of Bangladesh. American Eurasian Journal of Sustainable Agriculture 3(2):165-171.
|
|
|
|
|
Mishra MK, Nishani S, Gowda M, Padmajyothi D, Suresh N, Sreenath H, Raghuramulu Y (2014). Genetic Diversity Among Ethiopian Coffee (Coffea Arabica L.) Collections Available In Indian Gene Bank Using Sequence Related Amplified Polymorphism Markers. Plant Breeding and Seed Science 70(1):29-40.
Crossref
|
|
|
|
|
Murray MG, Thompson WF (1980). Rapid isolation of high molecular weight plant DNA. Nucleic Acids Research 8(19):4321-4326.
Crossref
|
|
|
|
|
Nadkarni KM, Nadkarni AK (1976). Indian MateriaMedica, Vol 1. 3rd ed. Mumbai, M/S Popular Prakasan Pvt. Ltd. http://krishikosh.egranth.ac.in
|
|
|
|
|
Nayampalli SS, Ainapure SS, Samant BD (1988). A comparative study of diuretic effects of Tinospora cordifolia and hydrochloro-thiazide in rats and a preliminary phase I study in human volunteers. Journal of Postgraduate Medicine 34(4):233-236.
|
|
|
|
|
Patel P, Rajkumar BK, Parmar P, Shah R, Krishnamurthy R (2018). Assessment of genetic diversity in Colletotrichum falcatum Went accessions based on RAPD and ISSR markers. Journal of Genetic Engineering and Biotechnology 16(1):153-159.
Crossref
|
|
|
|
|
Patel MN, Parmar LD, Parihar A, Singh AK, Sheikh WA (2014). A High- throughput DNA Extraction Protocol and its Utilization in Molecular Characterization of Noni (Morinda citrifolia L) Genotypes. Current Trends in Biotechnology and Pharmacy 8(2):166-174.
|
|
|
|
|
Padmalatha K, Prasad MN (2006). Genetic Diversity in Rauvolfia tetraphylla Lf using RAPD Markers. Journal of Plant Biotechnology 33(2):139-145.
Crossref
|
|
|
|
|
Padmalatha K, Prasad MN (2007). Inter and intrapopulation genetic diversity of Rauvolfia serpentina (L.) Benth. ex Kurz, an endangered medicinal plant, by RAPD analysis. Medicinal and Aromatic Plant Science and Biotechnology 1(1):118-123.
|
|
|
|
|
Pradeep Reddy, Sarla N, Siddiq EA (2002). Inter simple sequence repeat (ISSR) polymorphism and its application in plant breeding. Euphytica 128:9-17.
Crossref
|
|
|
|
|
Reddi KVNR, Krishnasatya A, Ramesh P, Prasad LS, Narendra K, Reddy PCOR, Reddy CVCM, Sekhar AC (2016). Molecular genetic assessment of Jatropha curcas L.germplasm of diverse origin along with its wild relatives for various for growth and early establishment
|
|
|
|
|
Raghunathan K, Mitra RN (1982). Pharmacognosy of indigenous drugs, central council for research in Ayurveda and Siddha. New Delhi 2:687-695.
|
|
|
|
|
Rana V, Thakur K, Sood R, Sharma V, Sharma TR (2012). Genetic diversity analysis of Tinospora cordifolia germplasm collected from northwestern Himalayan region of India. Journal of genetics 91(1):99-103.
Crossref
|
|
|
|
|
Reddy MP, Sarla N, Siddiq EA (2002). Inter simple sequence repeat (ISSR) polymorphism and its application in plant breeding. Euphytica 128(1):9-17.
Crossref
|
|
|
|
|
Ruan Y, Mo RH, Li M, Huang LQ, Luo Y, Li XY, Zhou J, Wu YS (2008). Molecular characters of Centella asiatica found with RAPD technology. Zhongyaocai Journal of Chinese medicinal materials 31(7):970-973.
|
|
|
|
|
Sambrook J, Fritsch EF, Maniatis T (1989). Molecular cloning: A laboratory manual. Cold Spring Harbor Laboratory Press.Cold Spring Harbor.New York. USA.
|
|
|
|
|
Shanti V, Nelson R (2013). Antibacterial activity of Tinospora cordifolia(Willd).Thoms on urinary tract pathogens. International Journal of Current Microbiology and Applied Sciences 2(6):190-194.
|
|
|
|
|
Sharaf-Eldin MA, Al-Tamimi A, Alam P, Elkholy SF, Jordan JR (2015). Genetic relatedness of artichoke (Cynara scolymus L.) hybrids using random amplified polymorphic DNA (RAPD) fingerprinting. Genetics and Molecular Research 14(4):18431-18439.
Crossref
|
|
|
|
|
Sharma SK, Dawson IK, Waugh R (1995). Relationship among cultivated and wild lentils revealed by RAPD analysis. Theoretical and Applied Genetics 91(4):647-654.
Crossref
|
|
|
|
|
Sivakumar V, Dhana Rajan MS, Mohamed Sadiq AM, Jayanthi M (2014). In vitro Micropropagation of Tinospora cordifolia (Willd.) Miers ex Hook. F. & Thoms - An Important Medicinal Plant. Journal of Pharmacognosy and Phytochemistry 3(2):5-10.
|
|
|
|
|
Singh DR, Shrawan S, Darick M, Anbananthan,V, Salim KM, Chaya K, Antia V (2012). Diversity of Morinda citrifolia L. in Andaman and Nicobar Islands (India) assessed through morphological and DNA markers. African Journal of Biotechnology 11(86):15214-15225.
|
|
|
|
|
Singh J, Sinha K, Sharma A, Mishra NP, Khanuja SP (2003). Traditional uses of Tinosporacordifolia(Guduchi). Journal of Medicinal and Aromatic Plant Science 25:748-751.
|
|
|
|
|
Tidke SA, Kiran S, Harke SN (2014). Analysis of genetic diversity in 20 cotton germplasm lines using Random Amplified Polymorphic DNA maker. Asian Journal of Plant Sciences 12(4-8):184-189.
Crossref
|
|
|
|
|
Tyagi P, Khanduja S, Kothari SL (2010). In vitro culture of Capparis decidua and assessment of clonal fidelity of the regenerated plants. Biologia plantarum 54(1):126-130.
Crossref
|
|
|
|
|
Wang S, Chen X, Han F, Li R, Li G, Zhao Y, Xu Y, Zhang L (2016). Genetic diversity and population structure of ginseng in China based on RAPD analysis. Open Life Sciences 11(1):387-390.
Crossref
|
|
|
|
|
Welsh J, McClelland M (1990). Fingerprinting genomes using PCR with arbitrary primers. Nucleic Acids Research 18(24):7213-7218.
Crossref
|
|
|
|
|
Yeh FC, Chong DK, Yang RC (1995). RAPD variation within and among natural populations of trembling aspen (Populustremuloide Michx.)from Alberta. Journal of Heredity 86(6):454-460.
Crossref
|
|
|
|
|
Yilmaz KU, Paydas-Kargi S, Dogan Y, Kafkas S (2012). Genetic diversity analysis based on ISSR, RAPD and SSR among Turkish Apricot Germplasms in Iran Caucasian eco-geographical group. Scientia Horticulturae 138(1):138-143.
Crossref
|
|
|
|
|
Zhang LJ, Dai SL (2010). Genetic variation within and among populations of Orychophragmus violaceus (Cruciferae) in China as detected by ISSR analysis. Genetic Resources and Crop Evolution 57(1):55-64.
Crossref
|
|
|
|
|
Zhao TF, Wang XK, Rimando AM, Che CT (1991). Folkloric medicinal plants: Tinospora sagittata var. cravaniana and Mahonia bealei. Planta Medica 57(5):505.
Crossref
|
|
|
|
|
Zhao Y, Chen XY, Wang XR, Pian RQ (2007). ISSR analysis of genetic diversity among Lespedeza bicolor populations. Journal of Plant Genetic Resources 8:195-199.
|
|
|
|
|
Zietkiewicz E, Rafalski A, Labuda D (1994). Genome fingerprinting by simple sequence repeat (SSR)-anchored polymerase chain reaction amplification. Genomics 20(2):176-83.
Crossref
|
|