2-C-Methyl-D-erythritol-4-phosphate (MEP) pathway has been extensively employed for terpenoids biosynthesis in Escherichia coli. In this study, to obtain key-enzymes of MEP pathway for squalene production, overexpression of different combination of MEP pathway genes were compared. Squalene production in strain YSS12 with overexpressed dxs, idi and ispA of MEP pathway from E. coli was improved by 71-fold when compared with strain YSS3 which only contained double copy SQS. Analysis of transcriptional levels of MEP pathway genes in engineering strains showed that different squalene production can be attributed to changed transcriptional levels of co-overexpressed genes dxs, idi, ispG and ispA in engineering strains. Furthermore, different E. coli expression hosts were compared for squalene production, among which BL21(DE3) was the best squalene producer. These results illustrate that dxs, idi and ispA of the MEP pathway from E. coli were key-enzymes for squalene production in E. coli. These key-enzymes of MEP pathway could also be applied to other terpenoids production in E. coli.
Squalene is a triterpene with a unique 30-carbon, polyunsaturated hydrocarbon and has a variety of pharmacological activities such as reduction of serum cholesterol levels (
Hien et al., 2017), anticancer (
Kotelevets et al., 2017), modulating fatty acid metabolism (
Kumar et al., 2016), and is extensively used in the functional food, cosmetic and pharmaceutical industries. Naturally, squalene is derived from two universal precursors, isopentenyl pyrophosphate (IPP) and dimethylallyl pyrophosphate (DMAPP), which are synthesized via the 2-C-methyl-D-erythritol-4-phosphate (MEP), or mevalonate (MVA) pathway (
Banerjee and Sharkey, 2014). IPP and DMAPP are condensed to form geranyl diphosphate (GPP) by FPP synthase, and subsequently condensed with another IPP to produce farnesyl diphosphate (FPP).
Finally, squalene is biosynthesized by a NADPH-mediated reaction catalyzed by squalene synthase (SQS) using FPP as the substrate (
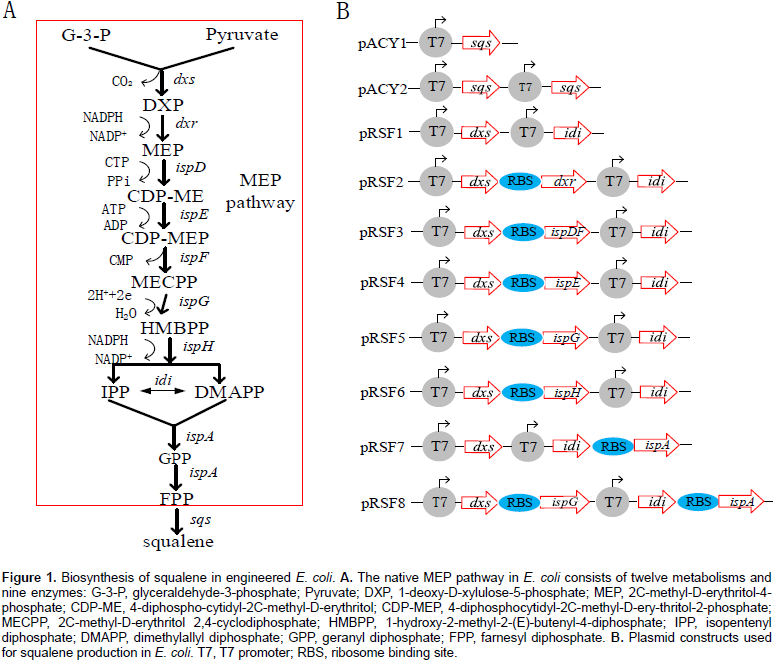
Ghimire et al., 2016)(Figure 1). MEP pathway is a natural metabolic pathway and only produce trace amount of IPP and DMAPP in
Escherichia coli that are precursors of all terpenoids. It consists of ten reactions catalyzed by nine enzymes (Figure 1a). Overexpression of MEP pathway genes were proven to be an effective method for increasing metabolic flux to IPP and DMAPP for terpenoid production in
Escherichia coli (
Jiang et al., 2012). DXS and IDI have been reported as the key-enzymes in the MEP pathway for increasing terpenoid production in
E. coli (
Yuan et al., 2006;
Zhao et al., 2013)and squalene production (
Ghimire et al., 2009).
Overexpression of genes
dxr (
Lv et al., 2016),
ispDF (
Ajikumar et al., 2010;
Yuan et al., 2006),
ispG (
Liu et al., 2014)and
ispA (
Han et al., 2016)were able to enhance terpenoids production. On the contrary, other studies have shown that overexpression of
dxr coupled with
dxs produced a similar isoprene level when compared with the
dxs overproduction strain (
Xue and Ahring, 2011), and overexpression of
ispDF together with
dxs and
idi resulted in decrease in terpenoids production (
Zhou et al., 2012). Meanwhile, genes
ispH,
ispE and
ispA have not been overexpressed in combination with
dxs and
idi genes of MEP pathway in the production of terpenoids in
E. coli. Based on the above studies, it is believed that in addition to the
dxs and
idi genes, other genes may be very important in the MEP pathway for squalene production in
E. coli.
In this study, in order to clarify key-enzymes of MEP pathway for squalene production in E. coli, squalene biosynthetic pathway was constructed by overexpressing SQS in E. coli. The authors also introduced different gene combinations of MEP pathway in the squalene producer to identify key-enzymes for squalene production and to study the correlation between the transcriptional levels of these genes and the yield of squalene. Finally, different E. coli strains were compared to determine the best host for squalene production.
Bacterial strains and culture conditions
All strains used in this study are listed in Table 1. E. coli DH5α were
Grown in LB medium at 37°C for plasmid construction.
E. coli BL21(DE3), BL21 Star(DE3), OverExpress C43(DE3) and Tuner(DE3) (Shanghai Weidi Biotechnology Co., Ltd) were used to produce squalene. Recombinant strains were cultured in fermentation medium (
Zheng et al., 2013)for squalene production. The cells were induced with 0.5 mM isopropyl β-D-thiogalactoside (IPTG) when OD
600 reached 0.6 to 0.9 at 30°C and 180 rpm for 48 h.
Construction of recombinant plasmids
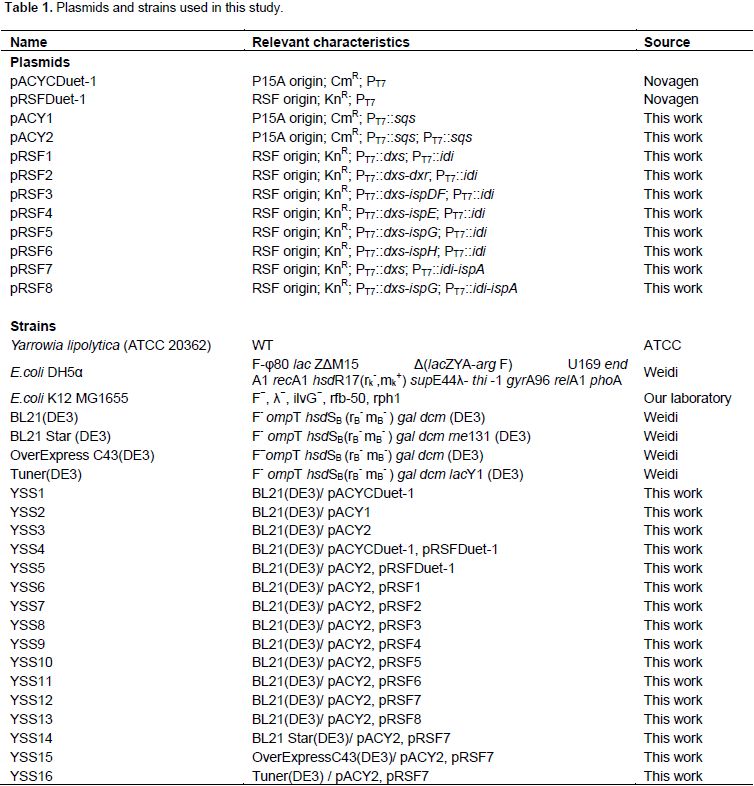
All plasmids used in this study are listed in Table 1 and all primers used in this study are listed in Supplementary Table S1. Molecular biology protocols were carried out as described in the literature (Sambrook and Russell, 2001). DNA fragments were amplified by polymerase chain reaction (PCR) using PrimeSTAR® Max DNA polymerase (TaKaRa, Dalian, China) according to the manufacturer’s instructions. All restriction enzymes and T4 DNA ligase were purchased from TakaRa (Dalian, China). DNA and plasmid extraction Kits were purchased from Shanghai Generay Biotech Co., Ltd. DNA sequencing and primers synthesis were provided by Shanghai Rui Di Biological Technology Co. Ltd. The gene sqs was PCR amplified from Yarrowia lipolytic and MEP pathway genes as well as fragments such as dxs-dxr, dxs-ispDF, dxs-ispE, dxs-ispG, dxs-ispH and idi-ispA involved in this study were PCR or overlap PCR amplified E. coli K12 MG1655 genomic DNA using corresponding primer set (Table S1). DNA fragments and vectors were excised with restriction enzymes (Table S1) and ligated with T4 DNA ligase to create corresponding plasmids (Table 1 and Figure 1B).

Identification and quantification of squalene
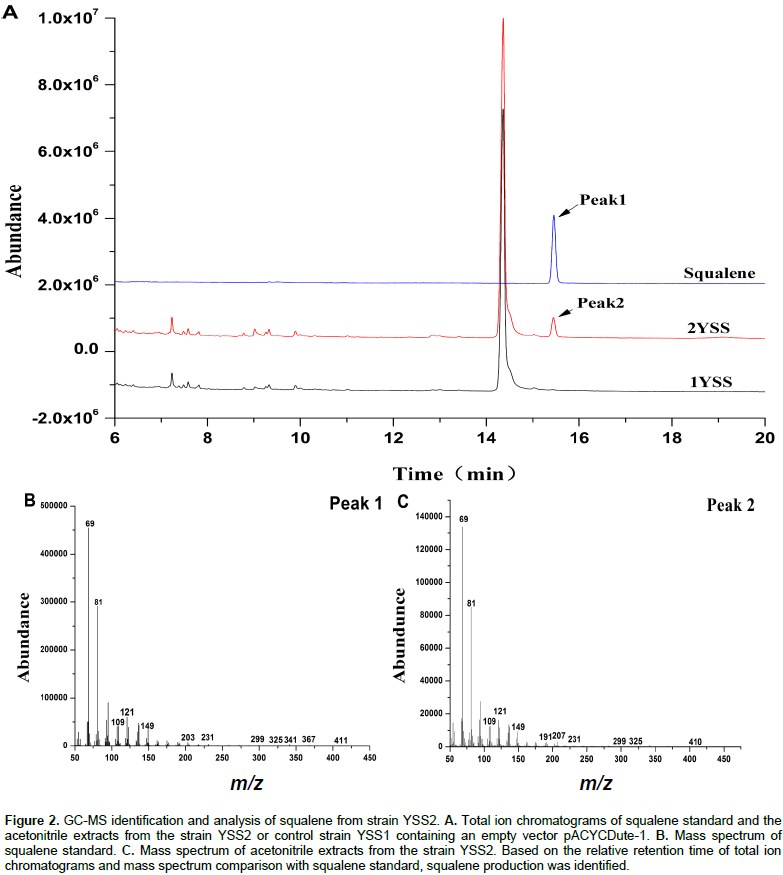
After centrifugation at 8000 rpm for 5 min, 20 mL culture medium were mixed gently with 10 mL hexane by inverting the tube 5 times. After another centrifugation at 8000 rpm for 5 min, the hexane phase was collected. This extraction process was repeated one more time. Meanwhile, cell pellets were disrupted by ultrasonic in 3 mL acetone for three times. Hexane and acetone extracts were combined and evaporated under reduced pressure. The dry residue was dissolved in 300 μL of acetonitrile and filtered through a 0.25 μm filter prior to GC-MS or HPLC quantitative analysis. The acetonitrile extracts (1 μL) were analyzed by GC-MS using a SHIMADZU GCMS-QP2010SE equipped with a Rxi-5ms (30 m × 0.25 mm × 0.25 µm) GC column. Compound separation was achieved with an injector temperature at 280°C, and a 30 min temperature gradient program for GC-separation starting at 200°C for 2 min followed by heating the column to 250°C at 20°C min-1 and a final constant hold at 250°C for 20.5 min.
Mass detection was achieved with electric ionization using an EI scan mode with diagnostic ion monitored: m/z 69, 81 and 149. Squalene purchased from Aladdin®, China was used as standard. For quantitative analysis of squalene, 20 μL acetonitrile extract was loaded onto an Agilent 1200 HPLC with UV detection at 210 nm. For chromatographic separation, a Waters SymmetryShieldTM RP18 column (250 mm × 4.6 mm, 5 μm) was used. The mobile phase consisted of 2% water and 98% acetonitrile. The solvent flow rate was 1.0 mL/min and the column was held at 40°C during the separation. The peak area was converted into squalene concentration according to a standard curve plotted with a set of known concentrations of squalene.
Quantitative RT-PCR analysis
Cells cultured under fermentation condition were harvested by centrifugation at 12000 rpm and 4°C for 1 min. Total RNA was isolated using TRIzol Reagent (Sangon, Shanghai) following the manufacturer’s instructions. RNA samples were treated with DNase I (TaKaRa, Dalian, China) for 30 min at 37°C. RNA was dissolved in 20 μL DEPC-H2O and stored at -80°C. cDNA was reverse-transcribed with Prime ScriptTM RT reagent Kit (TaKaRa, Dalian, China) following the manufacturer’s instruction and used as template for real-time PCR (qPCR). The primers used for qPCR are listed in Table S1. qPCR was carried out on a 7500 Real-Time PCR System (Applied Biosystems) using SYBR® Premix Ex TaqTM II kit (Tli RNaseH Plus) (TaKaRa, Dalian, China). The relative transcriptional levels were calculated by ΔΔCT method. The data were normalized using the clpB gene as an internal control. For each detected gene, the transcriptional level in control strain YSS4 was set to 1.
Establishment of squalene biosynthesis pathway in E. coli
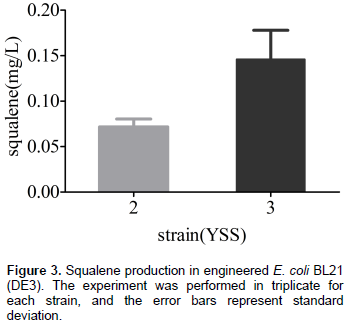
Although E. coli can synthesize FPP which is a precursor of squalene by a native MEP pathway, it is unable to produce the squalene because of the absence of SQS. In order to establish squalene synthetic pathway in E. coli (Figure 1), the gene sqs from Yarrowia lipolytica was sub-cloned into pACYCDuet-1 and transformed into E. coli BL21 (DE3) for the first time to get engineering strain YSS2. GC/MS analysis of cell extraction confirmed the presence of squalene (Figure 2). No squalene was detected in the control strain YSS1 that only harbors the vector pACYCDuet-1 (Figure 2). These results suggested that SQS from Y. lipolytica can be used for the biosynthesis of squalene in E. coli. Quantitative analysis of cell extraction by HPLC showed that YSS2 had a squalene yield of 0.072 mg/L at 48 h (Figure 3). The low production could be attributed to insufficient expression quantity of SQS and/or supply of precursors including IPP/DMAPP and FPP produced from native MEP pathway in E. coli.
The biosynthesis of squalene through overexpression of SQS in engineered E. coli
To increase the biosynthesis of terpenoid, it is also an effective way to overexpress the enzymes (Weaver et al., 2015). Trace amount of squalene produced by strain YSS2 may be because of low expression level of sqs. To enhance SQS concentration in E. coli, an extra copy of sqs was introduced into plasmid pACY1 resulting in the plasmid pACY2. The follow-up transformant YSS3 produced 0.15 mg/L squalene (Figure 3), which was approximately 2-fold of that produced by the strain YSS2. The results demonstrated that overexpression of sqs in E. coli is beneficial to the squalene production.
Finding key-enzymes of MEP pathway for squalene production in engineered E. coli
IPP and DMAPP are the universal precursors of all terpenoids in the living organisms (
Martin et al., 2003). Increasing cellular metabolic flux towards IPP and DMAPP is an effective strategy to improve yield of terpenoids production (
Leonard et al., 2010). In wild-type
E. coli, MEP pathway is the unique origin for providing IPP and DMAPP. Genes
dxs and
idi of MEP pathway have been widely engineered to enhance the supply of IPP and DMAPP concentration in
E. coli in order to increase synthesis of terpenoids (
Zhao et al., 2013). Thus,
dxs and
idi genes were cloned and introduced into YSS3, resulting in strain YSS6. According to the HPLC analysis, strain YSS6 produced 3.68 mg/L squalene (Figure 4), a 24-fold higher than the strain YSS3. The results demonstrated that the DXS and IDI are key-enzymes for squalene production in
E. coli. To determine whether other enzymes of MEP pathway affect the biosynthesis of squalene, the authors cloned
dxr,
ispDF,
ispE,
ispG,
ispH and
ispA genes from
E. coli K12 MG1655 genome to generate plasmids pRSF2, pRSF3, pRSF4, pRSF5, pRSF6 and pRSF7, respectively, as shown in Figure 1B. Co-transformation above plasmids respectively together with pACY2 resulted in six different strains.
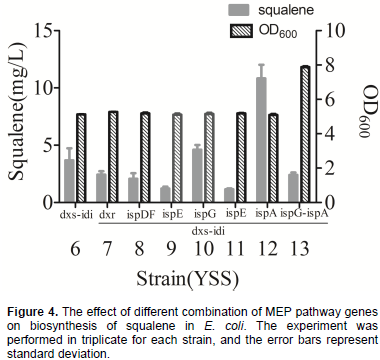
Figure 4 shows squalene production from these six different strains. Among these strain, YSS12 produced the highest squalene production at 10.83 mg/L, while YSS10 produced 4.60 mg/L, approximately 2-fold and 20% higher than that of YSS6, respectively. Squalene yields of strain YSS7, YSS8, YSS9 and YSS11 were 2.44, 2.07, 1.22 and 1.16 mg/L squalene, decreased by 34, 44, 67 and 68%, respectively, when compared with the YSS6. To further increase metabolic flux of the MEP pathway to squalene, plasmid pRSF8 was constructed by introducing dxs, ispG, idi and ispA, and together with plasmid pACY2, were co-transformed into BL21(DE3) to obtain strain YSS13. However, it only produced 1.6 mg/L squalene, decreased by 57, 65 and 85% when compared with YSS6, YSS10 and YSS12 (Figure 4). Comparing squalene production of all these strains, it showed that introducing dxs, idi and ispA could obtain highest squalene yield in YSS12 and the yield is approximately 71-fold when compared with the YSS3. Thus, DXS, IDI and IspA were considered to be key-enzyme of MEP pathway for squalene production in engineered E. coli.
Transcriptional levels analysis of MEP pathway genes in engineering E. coli
There are reports showing that metabolic imbalance by overexpression of certain genes in engineering metabolic pathway can lead to accumulation of toxic intermediates that produce inhibition of cell growth, metabolic flux overflow, gene transcription and enzymatic activity inhibition (
Kim and Copley, 2012). To illuminate the relationship of overexpression of genes of MEP pathway with varying squalene production in engineering
E. coli, the transcriptional levels of MEP pathway genes in these strains were measured by qPCR. As shown in Figure 5, when compared with the control strain YSS4, transcriptional levels of nine genes of MEP pathway were all weakly reduced in strain YSS5. This result illustrates that SQS/squalene could exert inhibitory effect on endogenous MEP pathway genes in engineering
E. coli. The transcriptional level of overexpressed genes of MEP pathway in corresponding strains was significantly increased; however, other non-overexpressed genes have no remarkable changes.
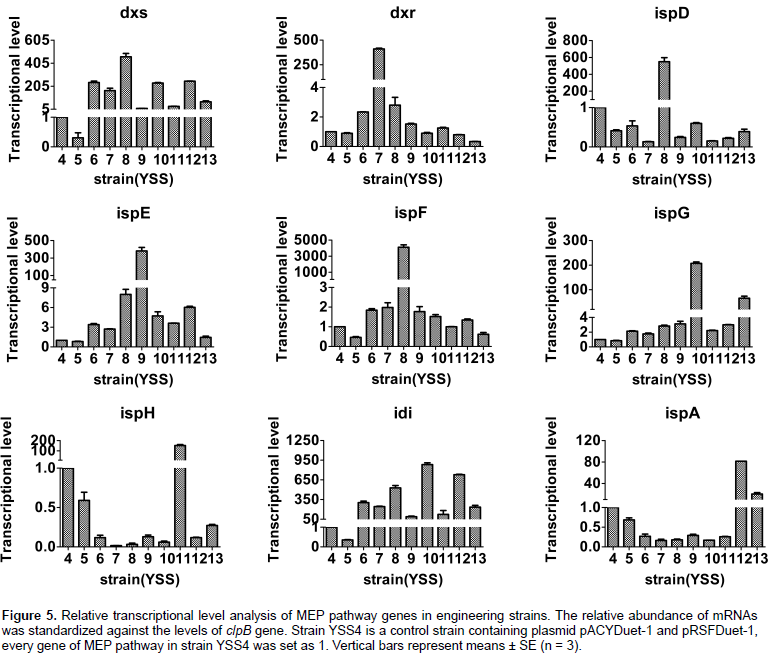
Compared with strain YSS6, transcriptional levels of dxs gene had about 36, 95 and 86% fold decrease in strains YSS7, 9, 11, and about 100, 0.5 and 7% increase in strain YSS8, 10 and 12. Transcriptional levels of idi gene had about 75, 193 and 143% increase in strains YSS8, 10 and 12 and about 19, 71 and 59% decrease in strains YSS7, 9 and 11. The transcriptional levels of dxs, ispG, idi, ispA genes in YSS13 were extremely reduced, as compared to YSS6, 10 and 12. Transcriptional levels of dxs and idi genes in YSS13 decreased by about 70 and 20% when compared with YSS6, and the transcriptional levels of ispG and ispA genes in YSS13 reduced by about 69 and 75% when compared with YSS10 and 12. These transcriptional results illustrated that overexpression of dxr, ispE, ispG, ispH and ispA and ispG genes of MEP pathway in strains 7, 9, 10, 11, 12 and 13 could influence transcriptional levels of co-overexpressed genes dxs, idi, ispG and ispA, which resulted in varying squalene production, except for the strain YSS8 with overexpression of dxs, idi and ispDF genes.
Comparison of squalene production in different E. coli strains
Metabolic pathway of terpenoids in
E. coli could be obviously influenced by host strain with different genetic background and lead to different terpenoids production (
Du et al., 2012). In order to choose an appropriate DE3
E. coli strain to maximize squalene production, pACY2 and pRSF7 plasmids were co-transformed into BL21 Star (DE3), OverExpress C43(DE3) and Tuner(DE3) strains, respectively to obtain strains YSS14, YSS15 and YSS16. After comparing these three strains with YSS12, it was found that YSS12 had the highest production of squalene (10.83 mg/L). YSS15 produced 9.28 mg/L squalene, YSS14 and YSS16 produced a much lower amount of squalene with 1.56 and 2.07 mg/L (Figure 6). Similar cell growth patterns were observed for all these strains. These results indicate that BL21 (DE3) was the most ideal strain for expression of the MEP pathway key enzyme DXS, IDI, IspA and SQS for squalene production.
In the process of terpenoid biosynthesis, introduction of exogenous MVA pathway into
E. coli resulted in successful improvement for terpenoid production (
Martin et al., 2003). However, previous study has demonstrated that native MEP pathway has a higher theoretical yield of terpenoid than MVA pathway in
E. coli by genome-scale
in silico modeling (
Meng et al., 2011). Katabami et al. (
2015)used truncated squalene synthases from human (
hsqs) in combination with MVA pathways to produce squalene up to 230 mg/L or 55 mg/g-DCW in flask culture, an approximately 55-fold increase as compared to
E. coli harboring
hsqs alone. In this study, in overexpression of key-enzymes genes
dxs,
idi and
ispA of MEP pathway with double copy of SQS from
Y. lipolytica in
E. coli BL21(DE3), yield of squalene increased 71 folds, when compared with the strain that only harbor two copies of SQS. The result once again proved that the MEP pathway is superior to the MVA pathway for terpenoids biosynthesis in
E. coli.
Presently, besides
dxs and
idi, other six genes of MEP pathway including
dxr (
Lv et al., 2016),
ispDF (
Ajikumar et al., 2010),
ispE (
Zhao et al., 2013)and
ispH (
Zhao et al., 2013)were explored for their potentials to increase terpenoids production. However, overexpression of
dxr,
ispDF,
ispH and
ispE together with
dxs and
idi decreased the squalene production when compared with the strain that overexpressed
dxs,
idi and double copy SQS, but enhanced squalene production when compared with the strain that only harbors two copies of SQS. These results illustrated that overexpression of
dxr,
ispDF,
ispH and
ispE coupled with
dxs and
idi did not substantially improve the yield of squalene but rather reduced DXS and IDI catalytic efficiency in the squalene production. Thus, it is suggested that overexpression of
dxr,
ispDF,
ispH and
ispE, respectively, together with
dxs and
idi are not helpful for terpenoids production in
E. coli.
The transcriptional levels of
dxs and
idi genes in strain YSS8 were higher than that in strain YSS6, but squalene production was lower. Previous report (
Zhou et al., 2012)showed that overexpression of
dxs,
idi and
ispDF could lead to over-production and accumulation of MECPP in cell to outflow into the broth that is toxic to MEP pathway, which further decreased the lycopene production. The same situation may also appear in the current study where the outflow of MECPP produced by overexpression of genes
dxs,
idi and
ispDF reduced toxic effect to MEP pathway, thus increased transcriptional levels of
dxs and
idi genes in strain YSS8. However, the outflow of MECPP also reduced the metabolic flux of the MEP pathway, and further decreased the production of squalene. IspG is a valuable enzyme in MEP pathway for terpenoids production (
Liu et al., 2014). In this study, similar result was observed by overexpression of
ispG together with
dxs and
idi genes.
However, squalene production was reduced by co-overexpression of dxs, ispG, idi and ispA in YSS13, and the transcriptional levels of overexpressed genes were also remarkably decreased when compared with strains YSS6, 10 and 12. This unexpected result may also be attributed to metabolic imbalance and toxic metabolites produced by overexpression of ispG together with dxs, idi and ispA in YSS13. Nevertheless, it is believed that IspG is an important enzyme and can be used to increase the production of squalene by balancing the flow of MEP pathway in future study.
IspA is also considered to be a key enzyme for terpenoids biosynthesis (
Han et al., 2016), however
ispA is not used extensively with MEP pathway genes but is widely used in the MVA pathway. Combination of
dxs,
idi and
ispA can increase squalene production in YSS12 by up to 71-fold when compared with the strain that only harbors two copies of SQS. This can be ascribed to a more balanced and productive MEP pathway metabolic flux to squalene by overexpression of
ispA. Therefore, it is suggested that overexpression of
dxs and
idi together with
ispA is an effective strategy for terpenoids biosynthesis in
E. coli.
In the engineering of E. coli that produced squalene of up to 0.15 mg/L, it was demonstrated that DXS, IDI and IspA of MEP pathway were key-enzymes for squalene production in BL21(DE3) by comparing the combinations of different MEP pathway genes. In brief, this work presented a promising strategy for the production of squalene in E. coli, and the key enzymes DXS, IDI and IspA could be used to effectively improve the production of other terpenoids in E. coli.
The authors have not declared any conflict of interests.