Backcross breeding enhanced by marker assisted selection is currently the most powerful method used to transfer one or a few genes controlling specific traits from one line into another. This method was used to cross naked barley variety, “Tombari” known as A, having high nutritional quality with hulled variety, “Giza131” called B, highly tolerant to drought. The objective of this work is to transfer the hulless variety into the drought tolerant variety. Five generations were realized. In each one, the best performing lines were chosen based on their agronomic, nutritional and molecular features. In the last generation, five remarkable barley lines were selected (H32, H33, H34, H36 and H47). In order to highlight the allele linked to the nudity trait, molecular profile of sKT7 marker, which is tightly linked to the “nud” locus, revealed two specific alleles (I and II). The first one (51 bp) was found in the two parental lines and their progenies. However, the second one (67 bp) was found only in the naked lines (parent A and progenies). This specific allele found only in the naked barley is different from that found by other researchers in Asia and North Europe. This means that naked barley has undergone multiple mutations. Some of the selected lines subjected to physiological stress induced by PEG-6000 showed greater tolerance to stress than their presumed tolerant parents.
Varietal selection has been in existence for a longtime where plants are selected according to some desirable traits with the best varieties or genotypes being crossed (Joshi and Witcombe, 1996). This selection has led to the diversification and creation of many improved varieties. To accomplish this objective, several breeding techniques were used differing in terms of efficiency and duration. The back cross method is one of these techniques. It is the most commonly used method to insert a trait into an elite cultivar (Bishwas et al., 2016). The variety that donates a gene is called “donor parent” while that which receives a gene is called “recipient parent”. The two
selected parents are crossed and hybrid is successively backcrossed to the recurrent parent. We introduce some interesting characteristics into the recurrent parent to improve one or two specific defects in it.
Backcrossing is used by many breeders for upgrading some inadequate traits within an existing elite cultivar. The characteristic could be a trait, a gene or even an anonymous locus or chromosome segment (Hospital, 2005). In each generation, progeny was chosen for the traits of interest and then backcrossed to the recurrent parent. In this scheme, the proportion of genome from the donor parent is reduced except for the part hosting the characteristic of interest. Usually, repeated backcrossing is mainly used to improve qualitatively inherited traits such as disease resistance, insect resistance and color, since the presence of target trait genes must be confirmed phenotypically at the individual level. Selection based on phenotypic level must clearly discriminate between segregating progeny. It has been used by breeders to distinguish between the recurrent parent and offspring (Yé et al., 2009; Hasan et al., 2015). Selection based on molecular marker alleles is mainly used when the effects of target alleles are difficult or expensive to measure phenotypically, and/or do not have a consistent phenotypic expression under certain specific selection conditions (Yé et al., 2009).
Several countries, particularly those in the North Africa, have their food ration based on barley. It is a species that adapts well to arid conditions, is rich in nutrients and some of its varieties are also rich in ß-glucan and proteins (Lahouar et al., 2017). Among the available barley varieties, some of them show great tolerance to drought. The use of these tolerant varieties in the production system would increase yield and yield stability in harsh environments while maintaining the same quality of harvested products. The combination of nutritional quality and drought tolerance represents one of the most complex challenges in the field of plant breeding. In recent years, incontestable research efforts have been made in genetics to identify the genes involved in nutritional quality (gene nud) and to understand the genetic determinism of tolerance to environmental stresses.
In this frame, the main objective of this work is to create new barley lines with tolerance to drought and having nutritional quality summarized in the traits coded by the gene (nud). In a previous work, we characterized sixteen North African barley genotypes based on their nutritional quality and drought tolerance levels. The quality characters are summed up in grain color, β-glucan and protein contents. Drought tolerance is based on yield traits under water deficit and some indices of drought tolerance. The results have led to the selection of a Tunisian naked genotype, with good nutritional quality (yellow grain color, high protein and ß-glucan content) called "Tombari" and an Egyptian drought tolerant genotype called "Giza131" (Ben Naceur et al., 2018, in press). Since the introgression of gene of interest is useful for genetic improvement in breeding programs, we have used these two genotypes in a scheme of back crossing process where “Tombari” genotype was used as donor parent and “Giza 131” as recurrent parent.
Plant material
Two barley genotypes (Hordium vulgare L.), "Tombari and G131" , with contrasting characters were crossed at the National Agronomic Research Institute of Tunisia. «Tombari or parent A» is a 6-row Tunisian landrace collected in 2000 from the South Tunisian region. It is a naked grain genotype, matures late with a prostrate habit, susceptible to disease and is characterized by an exceptional high nutritional quality. «Giza 131 or parent B» is a 6-row improved Egyptian genotype. It is vigorous with erected habit, matures half-early, is drought tolerant, resists fungal diseases and highly productive under favorable conditions.
Molecular analysis
The resulting F1 progeny (Tombari x G131) was controlled with molecular markers to check the crossing result. Then some of them were crossed with the recurrent parent (G131) to produce F2 or the first backcross generation (BC1F1). After molecular screening, the third (F3) generation was obtained by self-pollination to generate the BC1F2, on which we evaluate the agronomical traits and nutritional quality. The selected BC1F2 progenies based on agronomical and nutritional quality were crossed with the recurrent parent to produce the BC2F1 (F4 generation) which was self- pollinated to obtain BC2F2 (F5 generation). Selected plants are expected to be very similar to the recurrent parent and also possess the target traits (nutritional quality, precocity to escape terminal drought and tolerance to drought). Backcross progenies with the target traits were also selected based on molecular markers tightly linked to the genes controlling these traits (Table 1).
sKT7 is a dominant Sequence Characterized Amplified Region (SCAR) marker associated with the “nud” allele of the naked barley parent called “Kobinkatagi” (Taketa et al., 2004; Gustafsson, 2013). EBmac624 is a microsatellite region located close to the centromere of the chromosome 6H associated to the HvCO2 CO-like genes promoting flowering for barley (Wang et al., 2010).
Total DNA was extracted from young leaves of a single plant per genotype as described in Chaabane et al., (2012). The PCR reactions were carried out in a 25 μl reaction volume containing 18.8 µl ddH2O, 2 µl Taq Buffer 10x (containing MgCl2, 1.5 mM final concentration), 0.4 µl dNTP (10 mM), 0.8 µl forward primer (10 µM), 0.8 µl reverse primer (10 µM), 0.2 µl Taq polymerase (5 U/µl) and 2 µl template DNA (50 ng/µl). Amplifications were performed in a DNA thermocycler programmed for one cycle of 95°C for 2 min and 35 consecutive cycles of 1 min denaturing at 94°C, 1 min annealing at 55°C and 1 min elongation at 72°C followed by 2 min of post-elongation at 72°C. Amplified PCR products were separated by the “Experion” automated electrophoresis system (Bio-Rad). A 100-bp DNA ladder (Promega) was used as the molecular size standard.
Morphological and nutritional assessment
After molecular analysis, the plants having naked grain (“nud” marker allele) were evaluated for some morphological traits (main stem length, number of grains/spike, spike weight, 1000-kernel weight and grains yield) and nutritional quality (grain color, protein content and β-glucan content).
Root length assessment in stressful media
For each retained hybrid line and parent, 20 grains of barley were placed in 90-mm diameter Petri dishes lined with filter paper containing either distilled water (Control) or a polyethylene glycol solution (PEG-6000) at a concentration of 10% (Stress). Each treatment (stressed or none stressed) is repeated 4 times. The germination is carried out in a germination room (temperature: 25°C day/18°C night and 12 h light). After the incubation of the Petri dishes in the germination room for seven days, we measured the following parameters:
(i) The root length in the control and stressed conditions.
(ii) The stress tolerance index (STI) based on root length as defined by Fernandez (1992):
STI = [(Yp) x (Ys) /Ȳp2]
Where, Ys and Yp are the root length, respectively under stressed and non-stressed conditions, Ȳp is the mean length of all retained lines including the parental ones under favorable conditions.
Assessment of BC1 progeny (BC1F1)
Molecular analysis
To reduce the large number of donor chromosome, a minimum of six backcross generations is needed in the conventional backcrossing method. However, marker-assisted backcrossing may need two or three backcross generations only.
The crossing between A (Tombari) and B (Giza131) generated grains called F1. These grains were sown to verify if they were truly genetically different from the parental lines (A and B). Based on molecular test, only the F1 hybrid plants having both the marker alleles of the donor parent and recurrent parent were selected and backcrossed with the recurrent parent (Giza 131) to generate the first backcross generation (BC1F1). The molecular characterization by means of EBmac624 showed, in most cases, a new profile different from the parental lines (A or B). This means that the backcrossing between F1 and recurrent parent was successful. After elimination of the inbred lines, the rest of the offspring having the marker allele of the “nud” gene were labeled as H1, H2, H3, and H25. The barley seeds obtained during the BC1F1 were sown (auto crossed) to obtain BC1F2 (or F3 generation) for evaluation based on morphological and nutritional features.
Agro-morphological analysis
The BC1F1 progenies were sown for evaluation based on morphological and protein features and for implementing the second back-cross (BC2). At maturity stage, the main stems of each individual plant were measured along with the number of grain/spike, spike weight and 1000-kernel weight. Variance analysis of these parameters showed significant difference between the different progenies. The Newman-Keuls test allowed us to select the best hybrids (Table 2).
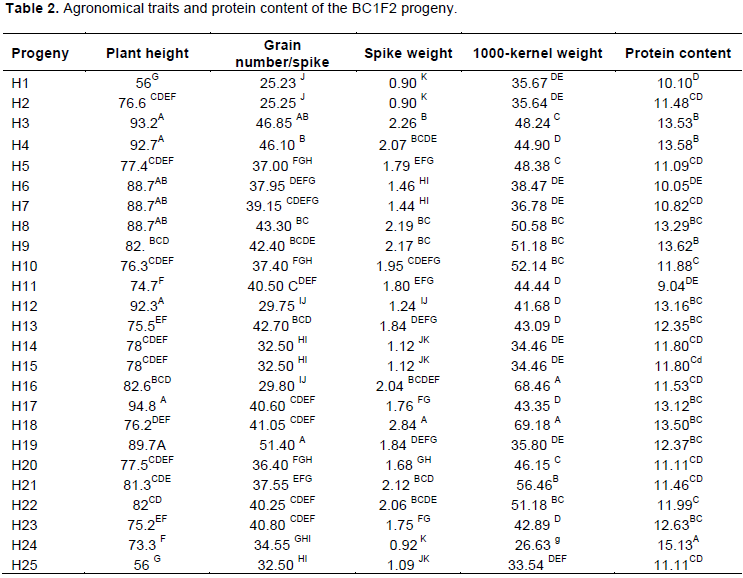
The hybrid plants having the longest stems (H3; H4 and H18), those having the highest main spike weight (H3; H8; H9 and H18), highest grain number/spike (H3; H4; H8; H9 and H18) and highest 1000-kernel weight (H3; H4; H8; H10; H16; H18; H22; H23) were selected. Among the analyzed hybrids, H3, H4, H8 and H18 seemed to be the most interesting plants.
Nutritional quality analysis
Protein analysis showed that H3, H4, H8, H9, H12, H17, H18 and H24 hybrids had the highest protein content (Table 2). The combination of those having the highest protein content and those having the best agronomic traits (H3, H4 H8 and H8 progenies) were selected and backcrossed with the recurrent parent (Giza 131) to obtain the BC2F1 progeny, which was auto crossed to obtain the BC2F2. The β-glucan content was not determined in this work, because we have shown in a previous work that the richness in β-glucan content correlated with the naked caryopsis (Ben Naceur, 2013). Therefore, from the beginning we have selected only the naked lines, and sure about their high β-glucan content.
Assessment of BC2F2 progeny
Agro-morphological characterization
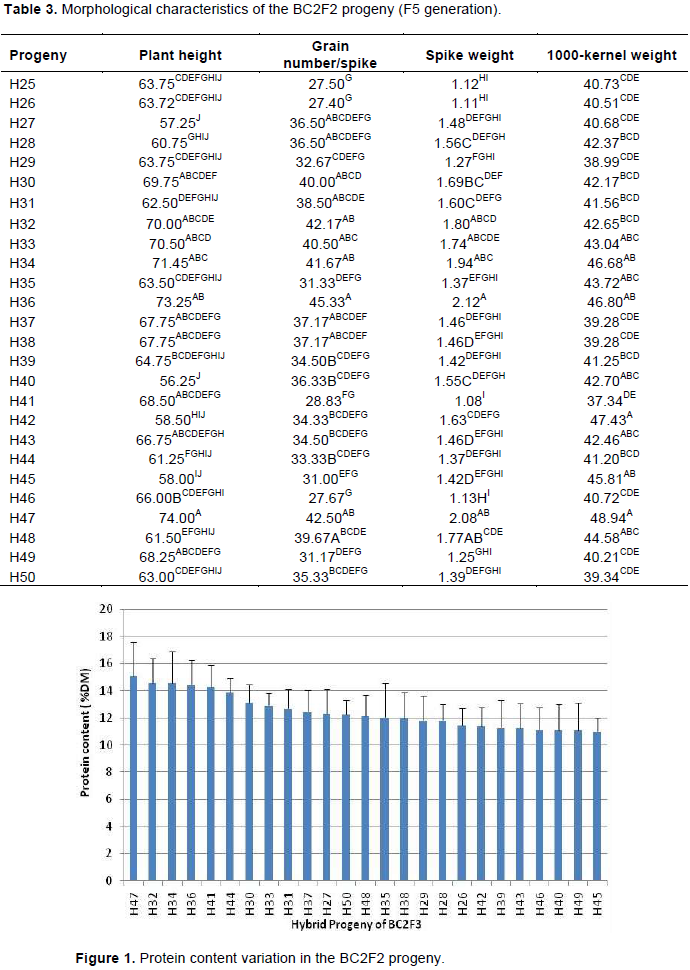
The BC2F2 seeds were sown and labeled as H26, H27, H28, H50. The morphological characterization is reported in Table 3. Analysis of variance revealed significant differences for all characters among the different BC2F2 progenies. The Newman-Keuls classification showed different separate classes for each characteristic. The class of longest plant height was composed of H30, H32, H33, H34, H36, H37, H38, H41, H43, H47 and H49, among which H32, H33, H34, H36 and H47 were the tallest. The class of highest number of grains/spike was composed of H32, H33, H34, H36 and H47. The class of highest spike weight was composed of H32, H34, H36, H47 and H48. Finally, the 1000-kernel weight showed the same trend as that of the spike weight and pointed out H32, H33, H34, H35, H36, H40, H42, H43, H45, H47 and H48 as the most important progenies, occupying the first class. Based on these characteristics, H32, H33, H34,
H36 and H47 were the most interesting hybrids.
Nutritional quality analysis (protein content)
Analysis of variance revealed significant differences (p<0.05) for protein content (Kjeldahl method) among the different BC2F2 progenies. The highest protein content was determined in H47; H32; H34; H36; H41 and H44 (Figure 1). In this group, the protein content varied between 13.87% (H44) and 15.11% (H47). The lowest content varied between 11% (H45) and 11.42% (H26). Such difference could be attributed to the interaction between the genetic background of the hybrid plants and the environment. The interaction (genotype x environment) affected barley protein content and was highly dependent on the cultivar as reported by several authors (Qi et al., 2006; Arendt and Zannini, 2013). Genotype–environment interactions for protein content in barley may also involve pleiotropic or linkage interactions with genes responsible for variation in these traits (Emebiri et al., 2005). Based on these two characterizations, the plants with the best agronomic and nutritional performance were H32, H33, H34, H36 and H47.
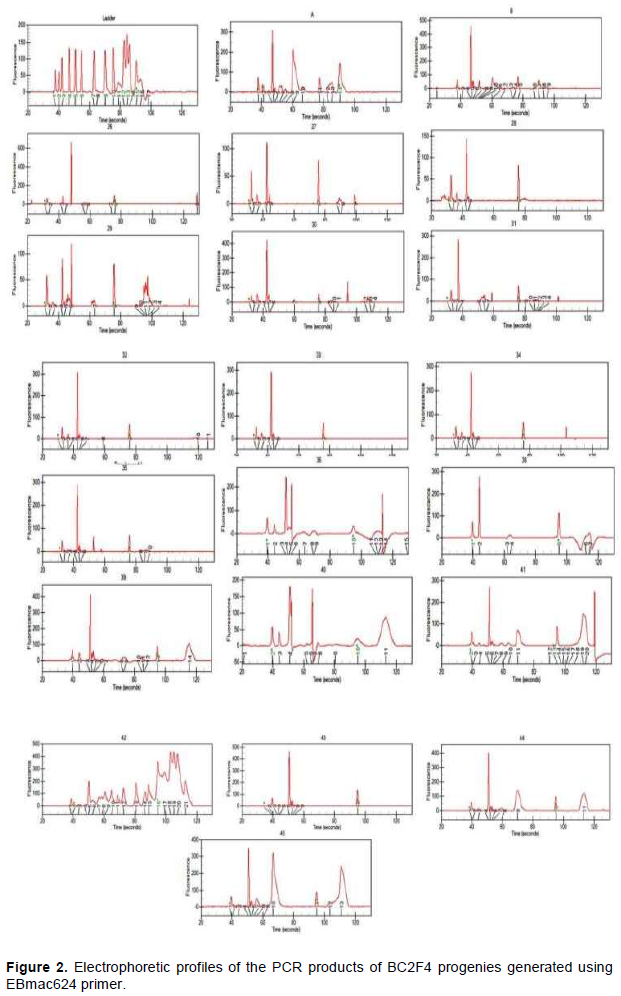
Molecular analysis
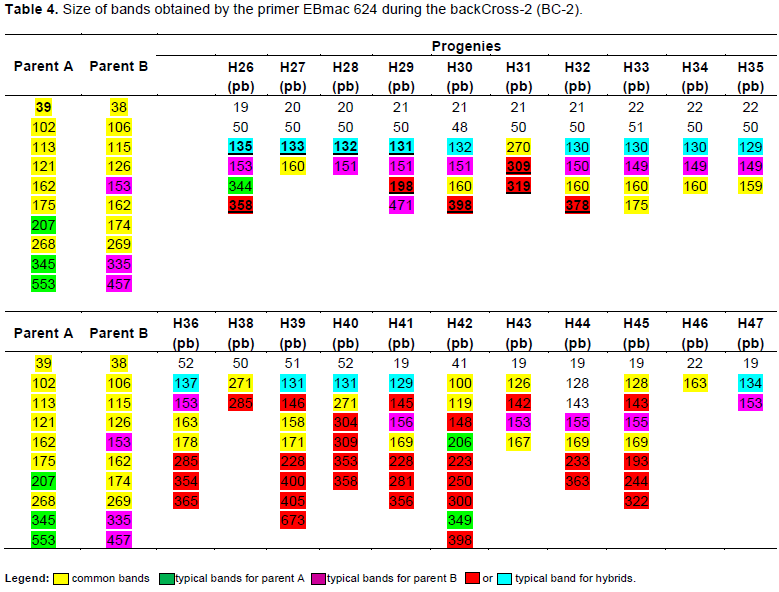
Electrophoresis profiles showing results of PCR products generated by EBmac624 primer are illustrated in Figure 2. Each peak materializes the band obtained during the separation of PCR products. Although some of them seem to be similar, all profiles are almost different from each other. Table 4 representing the band size confirms the difference among them. Marker assisted introgression is not always what can be guessed. Some crossing over might occur to generate new profiles different from the expected ones. According to Hospital (2005), many experiments have reported that genomic profiles of the produced progenies are relatively close to what is expected but not exactly identical to what is really obtained. In this case, the progenies carrying 151±2pb band are those having incorporated the genetic background of the recurrent parent (Giza131).
The result showed some progenies contain 131 bp bands (H26; H27; H28; H29; H30; H32; H33; H34; H35; H36; H39; H40; H41). Some others contain 151 bp bands (H26; H28; H29; H30; H32; H33; H34; H35; H36; H41; H43; H44; H45and H47) and some others contain either 131 or 151±2 bp band or both at the same time. Those containing the two bands are H26; H28; H29; H30; H32; H33; H34; H35; H36 and H41. According to this study, the progenies gathering the best agronomic performance and nutritional quality (naked), in addition to the genetic background of the parent B, are H32, H33, H34, H36 and H47 which would be evaluated as promoting lines or used in a third back-cross.
Confirmation of the “nud” gene presence in the selected hybrids
sKT is a sequence-characterized amplified region (SCAR) marker. It is the result of cross between two barley varieties: Kobinkatagi (naked barley) and Triumph (hulled barley). This marker co-segregates with the locus "nud" (Taketa et al., 2004). The PCR conducted on the hybrid DNA (H32, H33, H34, H36 and H47) and their parents (A and B) using the sKT7 primer generated the following profile (Figure 3) and the molecular weights associated with it (Table 5). The PCR product (Table 5) revealed two alleles: I and II. A 50bp allele was found in both parents and hybrids (except for H34) and a second allele of 67 bp was found only in naked barley (parent A and the naked progenies H32, H33, H34, H36 and H47). This allele (67 bp) could be associated with the “nud” gene in North African barley.
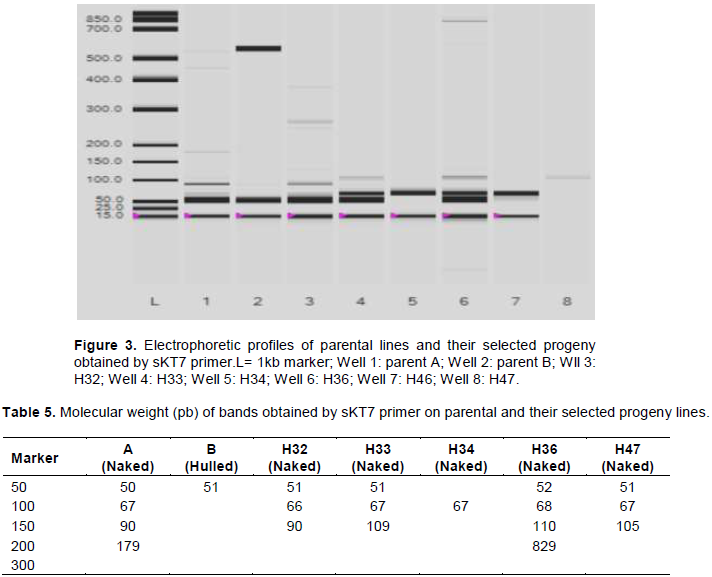
Taketa et al. (2004) worked on several accessions of Asiatic barley (wild, domesticated hulled and naked), using the same sKT7 primer; they identified a band of 470bp in their naked varieties. They concluded that the naked barley originated from a single mutation that occurred on the hulled barley in the Asian Region. In our opinion, the Taketa’s allele (470 bp) cannot be used as an indicator of all naked barley in the world. It would be specific for Asian barley only.
Furthermore, another study conducted by Saisho and Purugganan (2007) on naked barley from eastern and western Asia identified two closely related loci with different allelic frequencies. This suggests two independent origins of naked barley and contradicting the hypothesis of Taketa al. (2004) for which barley is controlled by a single locus, meaning a single origin. Archeological remains of naked barley have been found in the Nordic European countries (Finland), suggesting independent Finnish origin in naked barley. More recent work conducted by Gustafsson (2013) on Nordic barley (Northern Europe) using the same primer (sKT7) showed different profiles from what Taketa et al. (2004) found. This means that Nordic naked barley did not carry the same mutation reported by Taketa and suggests that naked barley is the result of several mutations occurring independently in Asia, North Europe and North Africa.
Effect of physiological stress induced by PEG-6000 on seedling growth
The ability of the plants to extend their root length in stressful conditions is a reliable indicator of the their tolerance to water deficit. This allows them to absorb water and nutrients in deep layers. That way we used the root elongation in stressful conditions to assess the selected barley lines’ tolerance to drought. Table 6 reveals the variability in root length both under favorable and stressful conditions induced by PEG-6000. Variance analysis of this parameter showed a significant difference (p<0.05) in both cases of water regime, indicating that the selected lines reacted differently to physiological stress. Under favorable conditions, the ANOVA and the Newman-Keuls classification showed 4 significantly different groups of which H36, H34 and H30 occupied the first group while H32 occupied the last group with an average varying between 3.2 and 7.7 cm. This result, which related to the genetic variability of the barley lines, is in agreement with those observed by Soltani et al. (2006) on wheat.
Under stress induced by PEG-6000, root length (RL) is greatly affected, but the response intensity and harmful effects of such stress depend on the genetic background of the lines. The highest average (RL) varied between 3.85 and 4.64 cm and was observed for the first group including H36, H47, H34, H32 and H33. On the other hand, the lowest (RL) was found at V10 line (1.82 cm). The ability of the first group, especially H36 line to develop their roots under water deficit conditions suggests the induction of certain genes involved in root elongation by stress or the structure modification allowing roots to sustain their development, in accordance with what was reported by Badiow et al. (2004).
The stress tolerance index (STI), based on root length, showed diversity among the lines in response to physiological drought (Figure 4). Assessing lines as per this selection index leads to the identification of few lines (H36 and H34) as drought tolerant ones. It showed also, the superiority of H36 compared to all other genotypes while V10 (parental line) appeared as the most sensitive. V30 (recurrent parental line) which is supposedly the most tolerant to drought appeared intermediate.