ABSTRACT
A new exotic weed, Flaveria bidentis, is spreading in central China where it forms dense monospecific patches modifying the structure of different native ecosystems and threatening native aboveground biodiversity. However, little is known about the consequences of such an invasion for soil bacterial community, especially its effect pattern at different invasion stages. In this study, soil samples were taken in native ecosystems that were uninvaded, partially invaded (transition), and severely invaded by F. bidentis. The bacterial richness and diversity in F. bidentis in rhizospheres soil was evaluated using denaturing gradient gel electrophoresis (DGGE) analysis. Different stages of F. bidentis invasion can trigger changes in soil physicochemical properties in particularly in available N and P F. bidentis invasion significantly decreased the richness of soil bacterial community, and the decline contents were positively correlated with invasion progress. In the invaded soils, bacterial species in Proteobacteria, Chloroflexi and Actinomycetes decreased with invasion, with the greatest reduction in relative abundance occurring for Proteobacteria, which was the dominant species in the native soils. Invasion of F. bidentis corresponded with an alteration in the structure of soil bacterial community, and soil microbial biomass as well, thus soil environment modification was expected to promote spreading of this exotic weeds in turn.
Key words: Biological invasion, Flaveria bidentis, soil nutrients, soil bacteria, polymerase chain reaction denaturing gradient gel electrophoresis (PCR-DGGE).
Abbreviation:
MBC, Microbial biomass carbon; MBN, microbial biomass nitrogen.
Biological invasion has become a global ecological and economic problem. Understanding the impact of invasive species on the local ecological systems has gained increased attention in invasive ecology (Ehrenfeld et al., 2001). Invasive species employs very complex and multiple mechanisms and strategies (Mack et al., 2000). Once establishing in a new environment, exotic weeds can replace the native species, causing above vegetable structural changes in the native ecological system. Soil microorganisms play an important role in the successful spread of exotic weeds (Hierro et al., 2005). Altered soil microbial communities and resulting effects on ecosystem processes may be an invisible legacy of exotic weed invasions by rhizosphere microbe and the host plants have an inter-dependent and mutual constraint relationship, which is the reason that why certain rhizosphere microbial communities only co-exist with specific plant species (Schloter, 2003). It has also been suggested that exotic weeds could disrupt mutualistic associations within native microbial communities (Richardson et al., 2000; Callaway and Ridenour, 2004; Stinson et al., 2006). Alteration in the structure and function of soil microbial communities could eventually lead to changes in the vegetation structures (Callaway et al., 2013). Among soil microbial communities, bacteria associated with plant roots are fundamentally important in plant nutrition, growth promotion, and disease interactions (Marschner et al., 2001). For this reason, there has been considerable interest in characterizing the structure and function of rhizosphere communities. The bacterial community composition in the rhizosphere is important for the performance of the plant, as bacterial species can have beneficial, neutral or harmful relationships with the roots (Atkinson and Watson, 2000; Sylvia and Chellemi, 2001). It is well accepted that different plant species can be selected for specific rhizosphere communities (Burke et al., 2002; Costa et al., 2006). The differences in root-derived substrates are claimed to explain the plant specific rhizosphere bacterial communities that have been observed for different plant species grown under similar conditions (Marschner et al., 2001; 2002; Smalla et al., 2001).
A new exotic weed, Flaveria bidentis (L.) Kuntze, commonly called “yellowtop”, is increasing spreading in central China. This species originated from South Africa and was first found in 2001 in suburbs of Tianjin and a few cities of Hebei province (Liu, 2005). It invades roadsides, abandoned field or even arable fields, out-competes natural vegetation, and forms a dense population (Huangfu et al., 2011). This weed tolerates environmental stresses of salinity and cold temperature, and could become troublesome for the development of sustainable agriculture (Gao, 2004). The function and population of rhizosphere microbes also undergo various alterations to allow the establishment of the invasive species. However, there is little information on the impact on soil bacterial diversity and mechanism upon invasion by the exotic weeds (Lorenzo et al., 2010).
This study aimed (1) to examine the effects of different stages of F. bidentis invasion on soil physicochemical properties and (2) detect the effects of different degrees of F. bidentis invasion on the community structure of bacteria in soils. We hypothesize that (1) increasing stages of F. bidentis invasion enhance soil nutrient element concentrations (especially soil N) because invasive plants have high nutrient cycling rates, especially for N (van Kleunen et al., 2010; Laungani and Knops, 2009; Jones and Chapman, 2011), and that (2) F. bidentis invasion significantly increases the richness and diversity of the soil bacterial community along the invasion gradients. Also, the changes in soil bacterial communities were associated with soil physicochemical properties. Towards these aims, we used the PCR- DGGE approach together with cloning and sequence analysis of 16S rRNA fragments of soil bacteria upon the invasion process by F. bidentis. In our findings will provide fundamental knowledge for soil bacteria diversity upon invasion by alien plant species.
Site description and sampling
The sampling sites were collected in wasteland ecosystems, a typical system F. bidentis infestation (Zhang et al., 2010), located in the Xian County in north China (38° 15′ 30″N, 115° 57′ 50″E) with temperate continental monsoon climate, mean annual precipitation of 560 mm, mean annual temperature of 12.3°C, and its average frostless period lasts 189 days. Geographically, the experimental site had flat land, a uniform landscape, and a similar terrain and soil origin with very minor disturbance by human and animals, and very minimal habitat variation. The soil is alluvial type where F. bidentis plants grown as monocultures had formed alternate successions. The following three sites (soil types) with three different levels of invasion by F. bidentis were sampled: (a) native soils (the control) mainly dominated by native herbaceous plants, including Setaria viridis (L.) Beauv, Digitaria ciliaris, Phragmites australis and Echinochloa crusgalli with coverage of over 60%; (b) transition soils where F. bidentis plants covered 10 to 30% of the plot, and (c) invaded soils where F. bidentis covered over 60% of the plot. Soil samples were collected on August 10 in 2009. In each of the sampling sites, six plots (repetitions) were randomly chosen each covering 3 × 3 m area with about 10 to 20 m apart from each other. The five points Quincunx sampling scheme was used to collect soil samples in each plots at the 0 to 20 cm depths, and soils within the same plot were pooled and mixed together equally as one replication, thus 18 soil samples collected in total were placed in plastic bags for transport to laboratory. Prior to sampling, all plants and organic matter debris on the ground were removed. Samples were stored at -20°C until analysis. From 1000 g of each soil sample, 20 g were homogenized and subsamples of 5 g were taken for further analysis. To verify the impact pattern found with this invasive plant, sampling was done in following year. Twice sampling data was pooled for soil physicochemical parameters analyses and only once PCR-DGGE fingerprinting was presented given the fact that there were no inter-year differences between treatments detected.
Determination of soil nutrients
Soil NH4+ and NO3- were extracted by shaking 20 g of fresh soil in 100 ml of 2 M KCl solution for 1 h. Soil extracts were analyzed with the FIAstar 5000 Auto Analyzer system. Total N and P in soil samples were analyzed with oven-dried samples, 48 h at 70°C. The Kjeldahl method was used for analyzing the total nitrogen (N) content of the soil. Soil mineral N was extracted using 2 mol L−1 KCl, then the concentrations of NO3–-N in the KCl extracts were determined by hydrazine sulfate colorimetry and the concentrations of NH4+-N by indophenol blue colorimetry (Mulvaney, 1996). Total phosphorus (P) was extracted using the HClO4-H2SO4 method, and available P was determined using the sodium bicarbonate method.
DNA extraction from soil samples
Total DNA was isolated from soil samples using the PowerSoil® DNA Isolation Kit (MO BIO Laboratories, Inc., CA), following the instruction of the maximum yield method. After a final purification, the soil DNA was visualized on 1% (w/v) agarose gels to assess its purity and molecular size. The final DNA extracts obtained from the soils were color-free, indicating that they did not contain high amounts of humic compounds.
Specific PCR of 16s rRNA gene fragments
For amplification of 16s rRNA fragments, a pair of universal primers consisting of the 357f- GC and 518r (Muyzer et al., 1993) were used to amplify the V3 region of bacterial 16S rRNA. Primer sequences were 357f- GC (5’-GCclamp-CCTACGGGAGGCAGCAG-3’), and 518r (5’-ATTACCGCG GCTGCTGG-3’). The PCR reaction were carried out in a final volume of 50 μl containing 2 μM of MgCl2, 200 μM of each dNTP, 0.5 μM of each primer, 50 ng of isolated DNA, 5 μl 10 × PCR buffer, and 2.5U of Ex TaqTM polymerase (TaKaRa Inc., Dalian, China). Touch-down PCR procedure was performed for increasing both the specificity and sensitivity of PCR assays in a thermal cycler (Bio-Rad) (Labbe et al., 2007). After preincubation at 95°C for 5 min, samples were amplified with denaturation for 1 min at 94°C, annealing for 1 min (temperature decreasing 0.5°C per cycle from 65 to 55°C, and then 15 cycles at 55°C), primer extension for 3 min at 72°C, followed by one final extension at 72°C for 15 min. Aliquots (5 μL) of PCR mixture were examined by electrophoresis in an agarose gel (1%, w/v) stained with ethidium bromide to check fragment size and integrity.
DGGE Patterns
DGGE was performed with 8%(w/v) acrylamide gels containing a linear chemical gradient ranging from 40 to 60%.The gels were allowed to polymerize overnight. DNA samples containing 20 μl of the PCR products were electrophoresed in 1× TAE buffer at 60°C at a constant voltage of 120 V for 8 h, and all DGGE analysis was done in the Dcode Universal Mutation Detection System (Bio-Rad, Hercules, CA, USA). After electrophoresis, the gels were stained for 30 min with SYBR gold nucleic acid gel stain (Invitrogen Molecular Probes, Eugene, USA) (10,000-fold diluted in 1× TAE) and photographed under UV light with a video imaging system. Band detection and quantification of band intensity was performed using Quantity One 4.62 software (Bio-Rad, USA). DNA band intensity was normalized by dividing the band intensity of each band by the mean band intensity of the gel. Therefore, both band position and intensity are expressed as relative values. Each peak represents individual groups of species having 16S rRNA sequences with similar melting behavior. The band intensity indicates the relative abundance of the group under these PCR conditions.
Sequence analysis of DGGE bands and phylogenetic analysis
Selected DGGE bands that occurred in majority of samples were excised from the gels and eluted (Kowalchuk et al., 1997). The criteria for selection of bands were that (i) they appeared as a single band in the pool of lanes, (ii) they represented bands in high abundance in the community or (iii) they were of relatively low abundance in the DGGE pattern. It is noteworthy that, in some cases where multiple clones were generated from a given excised band, more than one phylotype was detected from that band. In total, we sequenced 20 different clones, corresponding to 17 excised DNA fragments. These sequences of 16S rRNA genes obtained were submitted to the GenBank to determine the closest known relatives of the partial 16S rRNA sequences and the phylogenetic affiliations are shown in Table 2. Eluted DNA was then amplified using the 518r and 357f primer pair without GC clamp, and PCR products were ligated onto pMD19-T vector (Takara) and transferred into Escherichia coli JM109 competent cells. After positive cloning selection, the white colonies were further screened with vector primer pMD19-T to confirm the positive clones. The positive colonies were cultured in LB broth overnight at 37°C with constant shaking. Aliquots of 500 μL bacterial stocks were mixed with sterile glycerine (50%) and stored at -70°C. The clones of each of excised bands were chosen for sequencing. Sequencing was carried out at Shanghai Biotech Company. Searches in GenBank with the BLAST program (Altschul et al., 1997) were performed to determine the closest known relatives of the partial 16S rRNA sequences obtained. Multiple alignments of the sequences were performed using Clustal X (Thompson et al., 1997). A phylogenetic tree was constructed by the neighbor-joining method in MEGA 4.1 (Tamura et al., 2007). The confidence values for the branches of the phylogenetic tree were determined using bootstrap analysis (Felsenstein, 1985) based on 1000 resamplings. The similarity between sequences was calculated using the GENETYX computer program (Yumoto et al., 1999).
Measurement of soil microbial biomass carbon and nitrogen
Microbial biomass carbon (MBC) and nitrogen (MBN) were measured by a chloroform-fumigation extraction method modified from Vance et al. (1987). Six aliquots of wet soil from each sampling replications equal to 20 g dry weight were placed into 100 ml beakers. Three samples were fumigated whereas the other three were not. Soil was placed on top of the internal shelf in a vacuum desiccators drier that had internal diameter of 29 cm. Below the shelf, 60 ml HPLC grade chloroform and glass beads (to prevent explosion) were put into a 100 ml beaker. After addition of 50 ml 1 mol/L NaOH, the soil was covered with a few layers of wet filter paper to maintain moisture content of the steamed soil. After sealing the drier with Vaseline, vacuum was turned on and chloroform started to boil. Degas was topped after 5 min, and the samples were stored in darkness at 25°C for 24 h. After removal of the beaker containing chloroform, the soil was degassed again to remove chloroform residuals. The non-steaming treated soil was placed into a separate drier, and the chloroform was replaced with distilled water. After putting the fumigated soil into a 150 ml flask and addition of 60 ml 0.5 mol/l K2SO4 (soil: water = 1:4), the mixture was shaken at 25°C and 200 rpm for 30 min. The extracts were filtered through mid-speed filter paper, and the filtrates were measured immediately or stored at -15°C for later analysis. Soil microbial biomass carbon and nitrogen contents were measured using a multi NC3100 TOC/TN instrument (Analytik Jena AG, Germany). MBC and MBN of soil microbes were calculated using the differences in organic carbon and nitrogen between fumigated and non-fumigated soil, divided by the conversion factor of 0.45 (Joergensen, 1996). Data from the same soil type were pooled for analysis.
Statistical analysis
One-way ANOVA and Duncan's Test as post hoc test were used to check the differences in soil nutrients and microbial biomass C and N, bacterial richness and diversity between different soil types. Richness, defined as number of species, was calculated as the total number of bands per sample, and diversity, defined as number of different species and their relative frequency (Lorenzo et al., 2013). Based on DGGE results, cluster analysis of soil bacteria was performed using the Ward’s method by the Quantity-One software (Bio-Rad). The Shannon–Wiener index of general diversity, H was calculated using the following function:
H=−ΣPilog2 Pi (1)
Where, Pi is the importance probability of the bands in a track. H was calculated on the basis of the bands on the gel tracks using the intensity of the bands as judged by peak heights in the densitometric curves (Ampe and Miambi, 2000). The importance probability Pi was calculated as:
Pi=ni/Σn i (2)
ANOVA was performed with the SPSS 17.0 for Windows software package, and mean comparison was done using the least significant difference (LSD) test at P < 0.05.
Changes in soil nutrient contents status
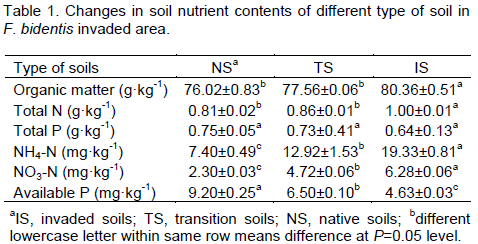
Compared with native soils, the F. bidentis invaded soils had obviously higher contents of soil organic carbon, total nitrogen, NO3-, and NH4+, but lower contents of soil available phosphorus (P<0.05). For example, it was increased by 5.7 and 23.4% in soil organic carbon and total nitrogen, respectively, while available phosphorous was reduced by 49.6% in invaded soil (P < 0.05, Table 1).
Impact of invasion by F. bidentis on bacteria diversity
16S rRNA fragments amplified from DNA extracted directly from soil samples were compared and only three replications of each site were presented due to a relatively high similarity of the DGGE patterns obtained for each of the replicates, which also suggested a low degree of variability caused by sampling (Figure 1). DGGE profiles of amplified 16S rRNA fragments from DNA extracted from the rhizosphere bacterial fractions revealed significant differences of the bacterial fingerprints from different F. bidentis invasion stages. Both the strains and number of bacterial reduced in well-invaded soils (Figure 1). Bands that were shared among all the soil samples included bands 2, 4, 6, 12 and 16, indicating that bacteria carrying these genes were common to all types of soil and were not affected by the invasion. The H’ were
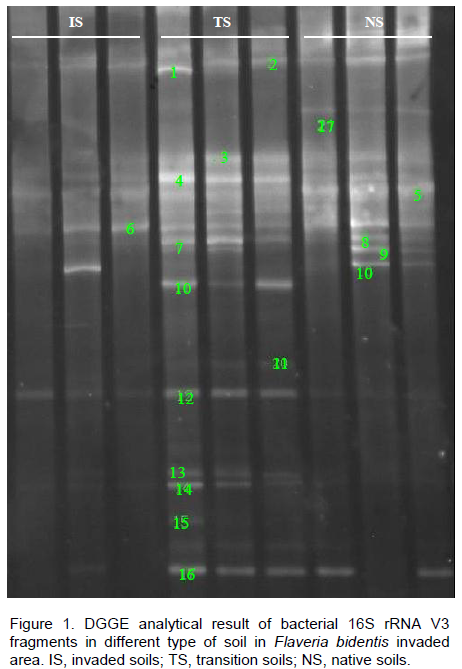
ranked in descending order as native soils (2.96) > transition soils (2.58) > invaded soils (2.33) (P < 0.05). Consequently, compared with the control (native soils), the invasion of F. bidentis reduced bacterial diversity with invasion progress.
Analysis of the DGGE profiles found were different in soil bacterial community between different soils. The transition soils and the native soils were clustered firstly at similarity index of 0.89 as one group, while the invaded soils distinctly separated from them with similarity of 0.68 (P < 0.05). The impact of this exotic weed on soil bacteria was a continuous process; as the invasion intensified some bacterial strains diminished in the soil. By comparing sequencing results, it was found that 20 sequences belonged to six different bacterial phyla with the majority in the division of Proteobacteria (Table 2). The similarity of the closest relatives of the partial 16S rRNA fragments of all sequenced bands ranged between 94 and 100%. Bands 2, 4-2, 7, 9, 10 and 16 showed the highest sequence similarity of 100%, respectively. On the other hand, other bands have various similarities with the
.png)
similar sequences in the NCBI database. The bands 1, 11 and 14 showed the higher sequence similarity (99%) with those of genus Kaistobacter, Rhodocyclaceae and Acidobacteriaceae, respectively, while bands 6, 8 and 13 only got 94% similarities with assigned sequences, respectively. According to the intensity of the band, a bacterial species that exhibited the higher sequence similarity to genus Kaistobacter (99% similarity) (band 1) was one of the most predominant during invasion of F. bidentis.
Impact of F. bidentis invasion on soil MBC and MBN
This result indicates that F. bidentis invasion led to alteration of microbial carbon metabolism in the soil (Figure 2). Upon the invasion of F. bidentis, soil MBC increased accordingly. The MBC content was ranked in descending order as following: invaded soils>transition soils> native soils. MBC was almostly 200% higher in the invaded soils than that of native soils (Figure 2a, P < 0.05). The same pattern was found for MBN which increased significantly after invasion by the exotic weed (Figure 2b). However, non-significant effect was detected between the transition soils and the native ones in MBN. Based on MBC and MBN, invasion of F. bidentis increased soil nutrient level as suggested in Table 1.
The results obtained partially support our original hypothesis. Firstly, invasion of F. bidentis lead to significant increases in soil N, organic matter, but decrease in available P. Because the genus of Flaveria is extensively cloned by arbuscular mycorrhizal fungi, the fungi known to help plant uptaking phosphorus from soils that are P-deficient for plant growth (Aziz et al., 1995; Bagayoko et al., 2000), further research should focus on possible competitive relationship in uptake of phosphorus between F. bidentis and native plant species. As a result, the decrease of phosphorus was attributed to the high uptake by this exotic weeds and the competition of exotic weeds with the soil community. Secondly, we found that invasion of F. bidentis was associated with significant increases in total soil N, C, organic matter and exchangeable P, but reduced soil bacterial diversity index, contrary to findings of Sanon et al. (2009). According to the positive feedback hypothesis, exotic weeds may cause soil-based ecosystem processes change following invasion, and such changes could establish positive feedbacks that enhance the spread of the exotic (Ehrenfeld et al., 2001). Our study suggests that number and diversity of soil bacteria changed, that the invasion of F. bidentis propagated certain groups of bacteria while suppressing others. Even some bacterial were common to all type soils, but plant species will eventually alter structure of soil bacterial community (Briones et al., 2002).
Many phytopathogenic organisms, bacteria as well as fungi, have coevolved with plants and show a high degree of host specificity (Raaijmakers et al., 2009). As the invasion progressed, the soil bacterial community structures also underwent significant changes as suggested by similarity analysis of the DGGE fingerprints where invaded soils clustered separately from native soils in the cluster analysis.
A decrease of bacterial diversity and increase of microbial biomass could be also caused by an increase of fungal biomass (Schimel et al., 1999). Alteration of native microbial community composition may further decrease competition from native plants and therefore support F. bidentis dominance as suggested by Rudgers and Orr (2009). F. bidentis can release allelopathic compounds (kaempferol, quercetin) (Xie et al., 2010; Iwashina, 2003), which may inhibit the growth of many microorganisms. It was found that extracts of F. bidentis from both leaves and roots reduced seed germination and seedling growth of native plant species (Huangfu et al., 2011). Therefore, allelopathic compounds produced by F. bidentis may be responsible for alterations in microbial biomass pools but further study is needed.
Our previous study has shown that F. bidentis invasion significantly decreases soil pH values (Zhang et al., 2010). This result may be mainly attributed to the fact that this invasive plant has high ammonium uptake rates as suggested by our study (unpublished data). The metabolic activities and community structure of soil microorganisms were highly correlated with soil pH values (Hackl et al., 2005; Högberg et al., 2007). Thus, we believe that changes in soil pH values mediated by F. bidentis invasion can enhance the succession of soil microbial communities in the rhizosphere and facilitate further invasion. With the widespread introduction and invasion of exotic weeds there are many studies that investigate alteration of basic ecosystem structure and function.
However, studies concerning invasive processes, information about changes in the impact over time is rarely available (Souza-Alonso et al., 2015). Some studies found that changes in soil properties as C or N contents and microbial properties soil ecosystem parameters are more pronounced after a long period of invasion (Marchante et al., 2008). Nevertheless, recent findings suggest that both ecological and adaptation processes may increase or attenuate the impact of invaders on the resident community, and that the impact of an invasive species on soil characteristics and on the structure and function of microorganisms does not necessarily remain constant or accumulate over the course of invasion (Strayer, 2012; Dostál et al., 2013). Our study sought to determine the effects of different stages of plant invasion on soil bacterial communities to better understand the mechanism of plant invasion. Different stages of F. bidentis invasion can trigger changes in soil physicochemical properties, in particularly in available N and P. F. bidentis invasion significantly decreased the richness of soil bacterial community, and the decline contents were positively correlated with invasion progress. Changes in the soil physicochemical properties and community structure of soil bacterial communities mediated by F. bidentis invasion may play an important role in facilitating further invasion.
The authors did not declare any conflict of interest.
This research was financially supported by the Natural Science Foundation of Tianjin (12JCQNJC09800), and Special Fund for Agro-scientific Research in the Public Interest (201103027).
MBC, Microbial biomass carbon; MBN, microbial biomass nitrogen.
REFERENCES
|
Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ (1997). Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402.
Crossref
|
|
|
|
Ampe F, Miambi E (2000). Cluster analysis, richness and biodiversity indexes derived from denaturing gradient gel electrophoresis fingerprints of bacterial communities demonstrate that traditional maize fermentations are driven by the transformation process. Int. J. Food Microbiol. 60:91-97.
Crossref
|
|
|
|
|
Atkinson D, Watson CA (2000). The beneficial rhizosphere: a dynamic entity. Appl. Soil Ecol. 15:99-104.
Crossref
|
|
|
|
|
Aziz T, Sylvia DM, Doren RF (1995). Activity and species composition of arbuscular mycorrhizal fungi following soil removal. Ecol. Appl. 5:776-784.
Crossref
|
|
|
|
|
Bagayoko M, George E, Romheld V, Buerkert A (2000). Effects of mycorrhizae and phosphorus on growth and nutrient uptake of millet, cowpea and sorghum on the West African soil. J. Agric. Sci. 135:399-407.
Crossref
|
|
|
|
|
Briones AM, Okabe S, Umemiya Y, Ramsig NB, Reichardt W, Okuyama H (2002). Influence of different cultivars on populations of ammonia-oxidizing bacteria in the root environment of rice. Appl. Environ. Microbiol. 68:3067-3075.
Crossref
|
|
|
|
|
Burke DJ, Hamerlynck EP, Hahn D (2002). Interactions among plant species and microorganisms in salt marsh sediments. Appl. Environ. Microbiol. 68: 1157–1164.
Crossref
|
|
|
|
|
Callaway RAM, Ontesinos DAM, Illiams KIW (2013). Native congeners provide biotic resistance to invasive Potentilla through soil biota. Ecology 94:1223–1229.
Crossref
|
|
|
|
|
Callaway RM, Ridenour WM (2004). Novel weapons: invasive success and the evolution of increased competitive ability. Front Ecol. Environ. 2: 436–443.
Crossref
|
|
|
|
|
Costa R, Götz M, Mrotzek N, Lottmann J, Berg G, Smalla K (2006). Effects of site and plant species on rhizosphere community structure as revealed by molecular analysis of microbial guilds. FEMS Microbiol. Ecol. 56: 236–249.
Crossref
|
|
|
|
|
Dostál P, Müllerová J, Pyšek P, Pergl J, Klinerová T (2013). The impact of an invasive plant changes over time. Ecol. Lett. 16: 1277-1284.
Crossref
|
|
|
|
|
Ehrenfeld JG, Kourtev P, Huang WZ (2001). Changes in soil functions following invasions of exotic understory plants in deciduous forests. Ecol. Appl. 11: 1287-1300.
Crossref
|
|
|
|
|
Felsenstein J (1985). Confidence limits on phylogenies: an approach using phylogenies: an approach using the bootstrap. Evolution 39: 783-791.
Crossref
|
|
|
|
|
Gao XM, Tang TG, Zheng LY, Zheng TX, Sang WG, Chen YL (2004). An alert regarding biological invasion by a new exotic plant, Flaveria bidentis, and strategies for its control. Biodivers. Sci. 12: 274-279.
|
|
|
|
|
Hackl E, Pfeffer M, Donat C, Bachmann G, Zechmeister-Boltenstern S (2005). Composition of the microbial communities in the mineral soil under different types of natural forest. Soil Biol. Biochem. 37: 661–671.
Crossref
|
|
|
|
|
Hierro JL, Maron JL, Callaway RM (2005). A biogeographical approach to plant invasions: the importance of studying exotics in their introduced and native range. J. Ecol. 93:5-15.
Crossref
|
|
|
|
|
Högberg MN, Högberg P, Myrold DD (2007). Is microbial community composition in boreal forest soils determined by pH, C-to-N ratio, the trees, or all three? Oecologia 150:590–601.
Crossref
|
|
|
|
|
Huangfu CH, Zhang TR, Chen DQ, Wang NN, Yang DL (2011). Assessing the residual effects of invasive weed Yellowtop for ecological restoration. Allelopathy J. 27:55-64.
|
|
|
|
|
Iwashina T (2003). Flavonoid function and activity to plants and other organisms. Biol. Sci. Space 17:24–44.
Crossref
|
|
|
|
|
Joergensen RG (1996). The fumigation-extraction method to estimate soil microbial biomass: calibration of the kEC factor. Soil Biol. Biochem. 28:33-37.
Crossref
|
|
|
|
|
Jones RO, Chapman SK (2011). The roles of biotic resistance and nitrogen deposition in regulating non-native understory plant diversity. Plant Soil 345:257-269.
Crossref
|
|
|
|
|
Kowalchuk GA, Gerards S, Woldendorp JW (1997). Detection and characterization of fungal infections of Ammophila arenaria (marram grass) roots by denaturing gradient gel electrophoresis of specifically amplified 18S rDNA. Appl. Environ. Microbiol. 63:3858-3865.
|
|
|
|
|
Labbe E, Lock L, Letamendia A, Gorska AE, Gryfe R, Gallinger S, Moses HL, Attisano L (2007). Transcriptional cooperation between the transforming growth factor-beta and Wnt pathways in mammary and intestinal tumorigenesis. Cancer Res. 67: 75–84.
Crossref
|
|
|
|
|
Laungani R, Knops JMH (2009). Species-driven changes in nitrogen cycling can provide a mechanism for plant invasions. Proc. Natl. Acad. Sci. USA 106: 12400–12405.
Crossref
|
|
|
|
|
Liu QR (2005). Flaveria Juss. (Compositae), a newly naturalized genus in China. Acta Phytotaxon Sin. 43: 178-180.
Crossref
|
|
|
|
|
Lorenzo P, Pereira CS, Rodríguez-Echeverría S. (2013). Differential impact on soil microbes of allelopathic compounds released by the invasive Acacia dealbata Link. Soil Biol. Biochem. 57: 156–163.
Crossref
|
|
|
|
|
Lorenzo P, Rodríguez-Echeverría S, González L, Freitas H (2010). Effect of invasive Acacia dealbata link on soil microorganisms as determined by PCR-DGGE. Appl. Soil Ecol. 44: 245–251.
Crossref
|
|
|
|
|
Mack RN, Simberloff D, Lonsdale WM, Evans H, Clout M, Bazzaz FA (2000). Biotic invasions: causes, epidemiology, global consequences, and control. Ecol. Appl. 10:689-710.
Crossref
|
|
|
|
|
Marchante E, Kjøller A, Struwe S, Freitas H (2008). Short- and long-term impacts of Acacia longifolia invasion on the belowground processes of a Mediterranean coastal dune ecosystem. Appl. Soil Ecol. 40:210-217.
Crossref
|
|
|
|
|
Marschner P, Neumann G, Kania A, Weiskopf L, Lieberei R (2002). Spatial and temporal dynamics of the microbial community structure in the rhizosphere of cluster roots of white lupin (Lupinus albus L.). Plant Soil 246:167-174.
Crossref
|
|
|
|
|
Marschner P, Yang CH, Lieberei R, Crowley DE (2001). Soil and plant specific effects on bacterial community composition in the rhizosphere. Soil Biol. Biochem. 33:1437-1445.
Crossref
|
|
|
|
|
Mulvaney RL (1996). Nitrogen-inorganic forms. In: Methods of Soil Analysis. Part 3, Chemical Methods. Madison, WI: SSSA.
|
|
|
|
|
Muyzer G, Waal EC, Uitterlinden AG (1993). Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction amplified genes coding for 16S rRNA. Appl. Environ. Microbiol. 59:695-700.
|
|
|
|
|
Raaijmakers JM, Paulitz CT, Steinberg C, Alabouvette C, Moënne-Loccoz Y (2009). The rhizosphere: a playground and battlefield for soil borne pathogens and beneficial microorganisms. Plant Soil 321:341-361.
Crossref
|
|
|
|
|
Richardson DM, Allsopp N, D'Antonio CM, Milton SJ, Rejmanek M (2000). Plant invasion – the role of mutualisms. Biol. Rev. 75:65–93.
Crossref
|
|
|
|
|
Rudgers JA, Orr SP (2009). Non-native grass alters growth of native tree species via leaf and soil microbes. J. Ecol. 97:247-255.
Crossref
|
|
|
|
|
Sanon A, Béguiristain T, Cébron A, Berthelin J, Ndoye I, Leyval C, Sylla S, Duponnois R (2009). Changes in soil diversity and global activities following invasions of the exotic invasive plant, Amaranthus viridis L., decrease the growth of native sahelian Acacia species. FEMS Microbiol. Ecol. 70: 118-131.
Crossref
|
|
|
|
|
Schimel JP, Gulledge JM, Clein-Curley JS, Lindstrom JE, Braddock JF (1999). Moisture effects on microbial activity and community structure in decomposing birch litter in the Alaskan taiga. Soil Biol. Biochem. 31:831-838.
Crossref
|
|
|
|
|
Schloter M, Bach HJ, Metz S, Sehy U, Munch JC (2003). Influence of precision farming on the microbial community structure and functions in nitrogen turnover. Agric. Ecosyst. Environ. 98:295-304.
Crossref
|
|
|
|
|
Smalla K, Wieland G, Buchner A, Zock A, Parzy J, Kaiser S, Roskot N, Heuer H, Berg G (2001). Bulk and rhizosphere soil bacterial communities studied by denaturing gradient gel electrophoresis: plant-dependent enrichment and seasonal shifts revealed. Appl. Environ. Microbiol. 67:4742-4751.
Crossref
|
|
|
|
|
Souza-Alonso P, Guisande-Collazo A, González L (2015). Gradualism in Acacia dealbata Link invasion: Impact on soil chemistry and microbial community over a chronological sequence. Soil Biol. Biochem. 80:315-323.
Crossref
|
|
|
|
|
Stinson KA, Campbell SA, Powell JR, Wolfe BE, Callaway RM, Thelen G, Hallett SG, Prati D, Klironomos JN (2006). Invasive plant suppresses the growth of native tree seedling by disrupting belowground mutualisms. Plos Biol. 4:727–731.
Crossref
|
|
|
|
|
Strayer DL (2012). Eight questions about invasions and ecosystem functioning. Ecol. Lett. 15:1199-1210.
Crossref
|
|
|
|
|
Sylvia DM, Chellemi DO (2001). Interactions among root-inhabiting fungi and their implications for biological control of root pathogens. Adv. Agron. 73:1-33.
Crossref
|
|
|
|
|
Tamura K, Dudley J, Nei M, Kumar S (2007). MEGA 4: Molecular evolutionary genetic analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24:1596-1599.
Crossref
|
|
|
|
|
Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997). The CLUSTAL X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nuclei Acids Res. 25:4876-4882.
Crossref
|
|
|
|
|
van Kleunen M, Weber E, Fischer M (2010). A meta-analysis of trait differences between invasive and non-invasive plant species. Ecol. Lett. 13:235-245.
Crossref
|
|
|
|
|
Vance ED, Brookes PC, Jenkinson DS (1987). An extraction method for measuring soil microbial biomass C. Soil Biol. Biochem. 19: 703-707.
Crossref
|
|
|
|
|
Xie QQ, Wei Y, Zhang GL (2010). Separation of flavonol glycosides from Flaveria bidentis (L.) Kuntze by high-speed counter-current chromatography. Sep. Purif. Technol. 72:229-233.
Crossref
|
|
|
|
|
Yumoto I, Iwata H, Sawabe T, Ueno K, Ichise N, Matsuyama H, Okuyama H, Kawasaki K (1999). Characterization of a facultatively psychrophilic bacterium, Vibrio rumoiensis sp. nov., that exhibits high catalase activity. Appl. Environ. Microbiol. 60:67-72.
|
|
|
|
|
Zhang TR, Huangfu CH, Bai XM, Yang DL (2010). Effects of Flaveria bidentis invasion on soil nutrient contents and enzyme activities. Chin. J. Ecol. 29(7):1353-1358.
|
|