ABSTRACT
A survey was conducted to determine the occurrence of sweet potato leaf curl virus (SPLCV) infecting sweet potato in Kenya. Vine cuttings of sweet potato were collected in 2011 from fields in Central, Western and Coastal regions of the country. The cuttings were grown in an insect-proof screen-house and scion grafted to Ipomoea setosa indicator plants. DNA was extracted from leaf samples of graft-inoculated I. serosa and tested for the presence of SPLCV by polymerase chain reaction (PCR) using degenerate and specific primers. The virus was detected in all the regions with incidence rates of 33, 17.0 and 2.6% from Western, Coastal and Central regions, respectively using specific primers. Coat protein gene AV1 from isolates collected from each region was sequenced and their comparison revealed >88.4% nucleotide identity. Phylogenetic grouping using nucleotide and amino acid sequences showed that four Kenyan isolates (Coast3, Central13, Coast15 and Western14) clustered together, while the other remaining five grouped a long with isolates from different parts of the world. The results reveal that Kenyan SPLCV isolates have a clear diversity and are also closely related with other isolates from different parts of the world. The wide distribution of the virus within the country means that urgent measures are needed to clean the farmers planting material to reduce possible losses incurred due to the presence of the virus.
Key words: Sweet potato leaf curl virus, coat protein gene, virus, sweet potato, nucleotide, amino acid.
Sweet potato (Ipomoea batatas (Lam) belongs to the family, Convolvulacea and is the third most important root crop after potato and cassava (Clark et al., 2012). Its energy-rich storage roots are consumed as a carbohydrate source and its leaves are used as vegetables (Antial et al., 2006). The crop is ranked fourth in importance in the developing world after rice, wheat and corn (Kays et al., 2005). Sweet potato is grown in all tropical or subtropical regions of the developed and developing world and is now the third most important root food crop globally after potato and cassava, largely in Asia and the Africa continents (FAOSTAT, 2015). Globally, the top five sweet potato producing nations at 2014 were China, Nigeria, Uganda, Indonesia and the United Republic of Tanzania (FAOSTAT, 2015). It is an important food security crop in Kenya, and the main production areas are Western, Nyanza, Central, Coastal and Eastern regions (Ateka et al., 2004). In Kenya, virus diseases cause major losses in sweet potato production (Njeru et al., 2004), the main diseases being Sweet potato virus disease (SPVD), a result of synergistic interaction between a potyvirus Sweet potato feathery mottle virus (SPFMV) and a crinivirus Sweet potato chlorotic stunt virus (SPCSV) (Ateka et al., 2004).
Over 20 different viruses belonging to at least nine families infect sweet potato worldwide (Clark et al., 2012; Valverde et al., 2007). Several of these viruses occur in East Africa, including SPFMV, (genus Potyvirus), Sweet potato mild mottle virus (SPMMV, genus Ipomovirus). Others includes those in the family Closteroviridae which, includes SPCSV (genus Crinivirus), Sweet potato chlorotic fleck virus (SPCFV, genus Carlavirus) and Sweet potato leaf curl virus (SPLCV, genus begomovirus) (Ateka et al., 2004; Miano et al., 2006; Wasswa et al., 2011). SPLCV is transmitted by Bemisia tabaci or through vegetative propagation. The virus also infects species of the Convolvulaceae which acts as alternative hosts after insect transmission (Valverde et al., 2003). SPLCV occurs worldwide (Briddon et al., et al 2006; Lotrakul et al., 1998). In Kenya, the virus was first reported in the Western region in a germplasm collection plot (Miano et al., 2006), but since then, no further work has been done to determine its overall distribution in the country. The aim of this study was to determine the distribution of the virus in the country and its genetic variability.
Sample collection
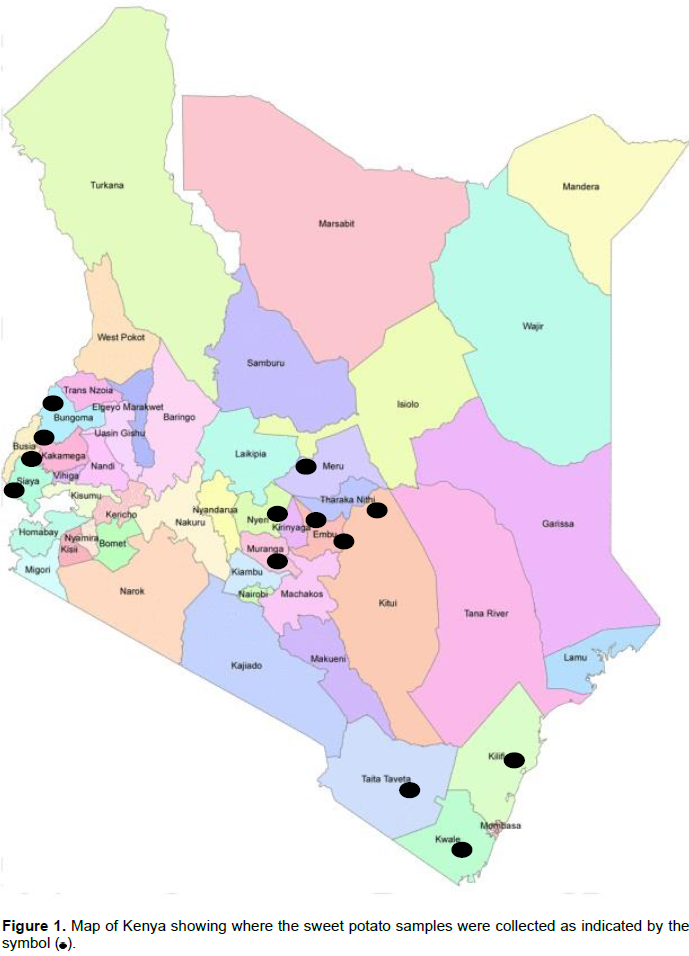
A survey was conducted between March and August 2011 in the major sweet potato growing regions of Kenya, namely Western (Kakamega, Bungoma, Busia and Siaya Counties), Coastal (Kilifi, Kwale and Taita Taveta Counties) and Central (Murang'a, Nyeri, Kirinyaga, Embu, Meru and Tharaka Nithi Counties) (Figure 1). Each field was selected at 10 km intervals along the major rural roads. A total of 512 samples were collected which included 97 from Western, 183 from Coast and 232 from Central region. Sweet potato vine cuttings with SPLCV like symptoms were collected, kept in separate polyethylene bags and transported in a cooler box to Biotechnology Centre, Kenya Agricultural and Livestock Research Organization (KARLO). Vines from each sample were then planted in plastic bags in sterile soil in an insect proof screen-house and symptoms that developed in the plants were recorded. Sweet potato cuttings were then grafted to Ipomoea setosa, a near-universal indicator host for sweet potato viruses and symptom development was observed for a period of four weeks. All plants were sprayed regularly with insecticides to control vectors.
DNA extraction
Samples for DNA extraction were collected by taking symptomatic leaf samples from graft inoculated I. setosa plants. DNA was extracted from the leaves using cetyl trimethyl ammonium bromide (CTAB) extraction method (Clarke et al., 2009; Li et al 2004). Polymerase chain reaction was done using a specific primer pair for SPLCV (SPB1/2) and degenerate primer pair (SPG1/2) for general Begomoviruses (Li et al., 2004). The specific primers SPB1/2 were used to amplify the coat protein (CP) gene (AV1) while the degenerate primers amplified AC1 gene. All amplifications were performed on a Gene Amp PCR system 9700 (Applied Biosystems) in 25-μl reaction mixtures containing 2 μl of the DNA extract, 0.5 μl of each primer (10 μM), 0.5 μl of 10 mM dNTP mix, 1.6 μl of Taq DNA polymerase (BIOLABS USA), 2.5 μl reaction buffer, 2.5 μl of 25 mM MgCl2 and 14.9 μl of molecular grade water. PCR conditions used were as follows: 94°C for 3 min, 35 cycles of (94°C for 1 min, 55°C for 1 min and 72°C for 2 min) and 72°C for 10 min. PCR products were visualized using electrophoresis in 2% agarose gel under ultraviolet illuminator after staining with gel redTM. PCR products from nine isolates were selected and purified using Qiagen purification kit according to the manufacturer's instructions and sent for sequencing to Macrogen, Amsterdam for Sanger sequencing.
Coat protein sequence analysis
The nucleotide sequences obtained from the nine selected isolates were compared with other ones in GenBank. The DNA sequences were aligned and calculation of sequence identity was done using Clustal Omega. Dendrogram were constructed using the neighbor-joining method supported with a bootstrap test of 1000 replicates in MEGA 6 using Tamura-Nei model method (Tamura et al., 2013). The nucleotide sequences described in this study were deposited in GenBank under accession numbers, KJ844869-KJ844877.
Incidence of Sweet potato leaf curl virus in Kenya
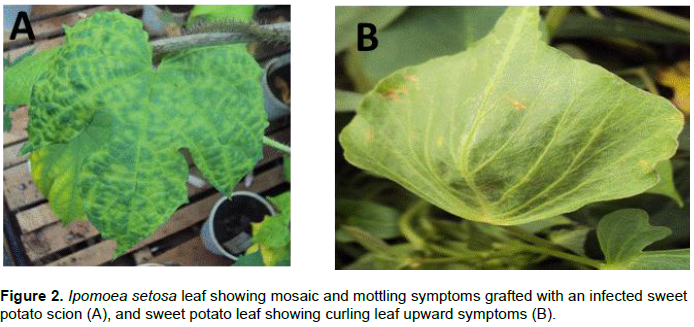
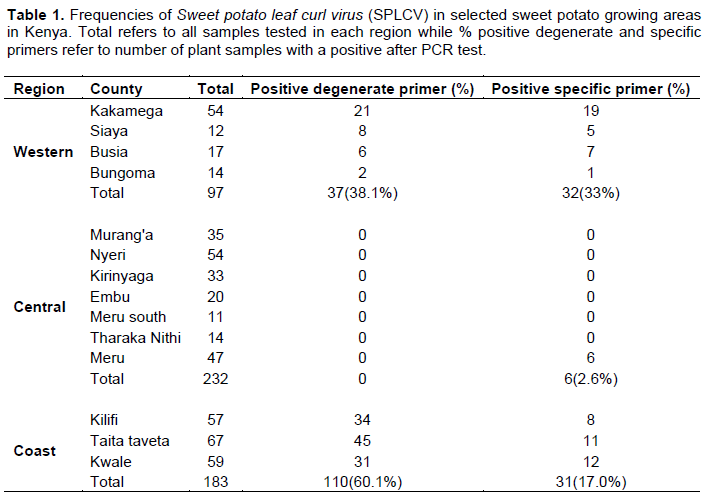
A total of 512 samples were collected in Western (97), Coast (183) and Central (232) regions with most of them showing virus-like field symptoms which included mosaic, curly leaf, yellowing, vein clearing, stunting, upward cupping, reduced leaf size and chlorosis (Figure 2A and B). After grafting, most of the symptoms from the field were still observed in host indicator plant I. setosa (Figure 2B). Out of the 97 samples collected from the Western region, 32 (33%) and 37 (38.1%) were positive when tested using specific and degenerate primers, respectively (Table 1). The incidence with specific primer by county was Siaya at 41.7%, Busia 41.2%, Kakamega 35.2% and Bungoma 7.1%. In Central region, 232 samples were collected and only six (2.6%) tested positive for SPLCV. The six positive samples were all from Meru County. There was no other sample from that Central region that was positive, using degenerate primer. Out of the 183 samples collected and tested from Coast region, only 31 samples (17%) were positive using specific primer, while 110 samples (60.1%) were positive using degenerate primers. Within the Coast region, SPLCV incidence in Kwale was 20.3%, Taita Taveta 16.4% and Kilifi 14%. Using degenerate primers, the incidence of begomoviruses in the Coast region was higher than in the Western region. Within the Coast region, Taita Taveta County had the highest incidence of begomoviruses (67.2%), followed closely by Kilifi (59.6%) and Kwale (52.5%). High begomoviruses incidence was reported in Siaya County (66.7%) in Western regionas compared to Kakamega (38.9%), Busia (35.3%) and Bungoma (14.3%). The nine samples yielded sufficient product for further molecular characterization.
Molecular analysis of partial AV1 coat protein gene
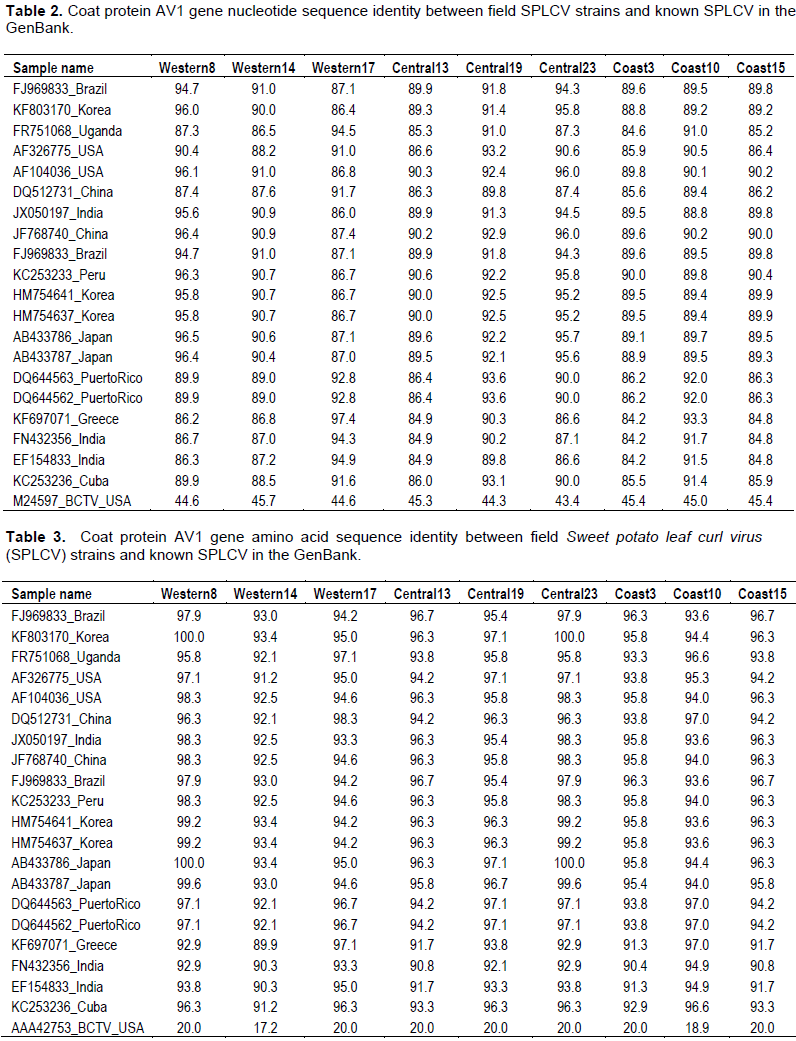
Partial coat protein gene (AV1) sequences of nine SPLCV isolates from Kenya had nucleotide identity (nt) and amino acid sequence identity (aa) of 84.6 to 99.9 and 91.2 to 100%, respectively (Tables 2 and 3). The phylogenetic analysis also showed that the Kenyan isolates are highly diverse with some isolates clustering in a single branch (Figures 3 and 4). When sequences of SPLCV isolates from Kenya were aligned against known isolates from the GenBank, they had nt and aa sequence identity ranging from 84.2 to 97.4% and 89 to 100%, respectively. The isolates from Western and Central Kenya had 100% aa sequence similarities as compared to isolates from Korea and Japan. The phylogenetic analysis clustered Kenyan isolates into 3 clusters; one cluster being unique only to Kenyan isolates (Figures 3 and 4). The SPLCV isolates from the Coast region had aa sequence similarity of 96.7 to 97% as compared to those from Puerto Rico and Brazil. The phylogenetic analysis based on nucleotide sequences showed two major clusters with each branching being supported by over 97% bootstrap value. Kenya SPLCV isolates were found in both clusters. Isolate Western17 grouped very differently when compared with other isolates from Kenya but grouped together with Ugandan isolate. Four Kenyan strains grouped together (Figure 1) in one of the major clusters.
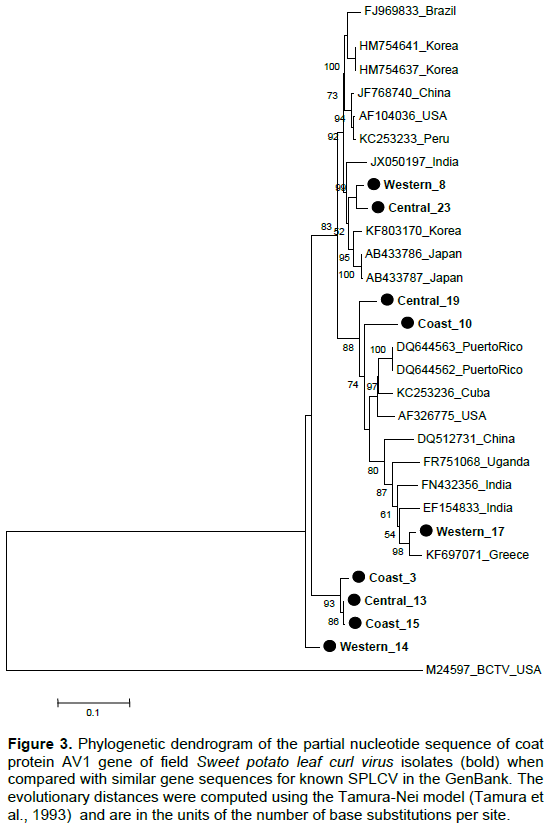

This research reports for the first time, the presence of Sweet potato leaf curl virus in all sweet potato growing regions of Kenya with Siaya and Busia having the highest incidences of 41.7 and 41.2%, respectively. Central region had some counties with no incidence reported at all, Bungoma with 7.1% had the lowest incidences of SPLCV reported in Western Kenya which was significantly lower than 35.2% found in Kakamega County. In the Central region, incidence of SPLCV was lowest in the countrywide since out of 232 samples collected, only six or 2.6% were positive. In the Coastal region, incidence of SPLCV was moderate, ranging from 14% in Kilifi to 20.3% in Kwale. Among the regions, Western Kenya had the highest risk of SPLCV followed by Coast region and Central region with the lowest.
The results obtained in this study reported the presence of SPLCV in Western Kenya, in agreement with previous studies that SPLCV was present in the region and they are (Miano et al., 2006). The presence and high
incidence of the virus in the Coast region was not expected. Degenerate primer used for begomoviruses in sweet potato detected 60.1% of viruses. This was significantly higher than 17.0% detected using specific primer. This could be due to its universal broad detection (Li et al., 2013). The presence of SPLCV in Taita Taveta could be attributed to cross-border trade with Tanzania where begomoviruses have been recently reported (Mbanzibwa et al., 2014). The Central region had the least SPLCV incidence. One of the reason could be the geographical elevation where the highlands have cool and wet conditions (Thornton et al., 2009), which may not be favorable for vectors (Thresh et al., 2003). The other reason could be the mode of cultivation as compared to Western region where sweet potato is grown throughout the year. The economic importance of sweet potato begomoviruses in Kenya could have been overlooked, probably because sometimes when some genotypes are infected with SPLCV, they do not show symptoms, and when present, they are not common and do not persist in sweet potato or may be difficult to notice (Valverde et al., 2007). This may be one of the reasons why the virus is widely distributed in most regions of the country. In addition, movement of the infected storage tubers could also introduce viruses within the small holder farms, as storage roots have been reported to spread the viruses through vegetative propagation (Okada et al., 2017).
Coat protein genes have been used in taxonomy of begomoviruses (Padidam et al., 1999) with the nucleotide and amino acid cutoff score of <89 and <90% respectively. Nine viral isolates were sequenced, three from Western, three from Coast and three from Central region. The genetic analysis of the partial AV1 gene sequence revealed that Kenyan SPLCV isolates are highly diverse, though some of the isolates detected in Western were closely related to those in Coast and Central regions. This could be attributed to movement of people carrying planting materials from one region to another. The phylogenetic analysis showed two major groups of SPLCV with Kenyan isolates being distributed within the two groups. Previous studies have reported that AV1 gene is the most similar gene in begomoviruses of the same geographical area (Harrison and Robinson, 1999). However, this was different from Kenyan isolates since those from the coast region were closely related to isolates detected in the South America (Puerto Rico and Brazil), which could be attributed to the fact that some sweet potato genotypes grown in Kenya originated from South America (O’brien, 1972), while those detected in the Central and Western regions were closely related to isolates from Asia (Korea, China and Japan) an indication that the Kenyan strains might have multiple origin (Luan et al., 2007).
In conclusion, a high incidence of SPLCV was found in sweet potato fields in Western and Coastal regions of Kenya. Since SPLCV could be associated with major losses, the two areas need to be given priority when providing farmers with virus-tested sweet potato planting materials. There is also need to minimize movement of virus-infected sweet potato materials from highly infected areas of Coastal and Western parts of the country to other places like Central region where the prevalence is low. Since SPLCV and other viruses are common in sweet potato growing areas of Kenya, there is need to study whether synergistic interactions or co-infections with other viruses occur which may result in increased losses. It will be important to follow up the same research in Kenya with current advent of genomics (Maina et al., 2016), to reveal other present sweet potato viruses that may not be detected with specific traditional diagnostics that may pose threat to sweet potato production. Future research should also involve whole genome genetic recombination studies to establish if virus recombination has already taken place and its amplifications.
The authors did not declare any conflict of interest.
REFERENCES
|
Antial BS, Akpanz EJ, Okonl PA, Umorenl IU (2006). Nutritive and Anti-Nutritive Evaluation of Sweet potatoes. Pak. J. Nutr. 5(2):166-168.
Crossref
|
|
|
|
Ateka EM, Njeru RW, Kibaru AG, Kimenju JW, Barg E, Gibson RW, Vetten HJ (2004). Identification and distribution of viruses infecting sweet potato in Kenya. Ann. Appl. Biol. 144(3):371-379.
Crossref
|
|
|
|
|
Briddon RW, Bull SE, Bedford ID (2006). Occurrence of Sweet potato leaf curl virus in Sicily. Plant. Pathol. 55(2):286-286.
Crossref
|
|
|
|
|
Clark CA, Davis JA, Abad JA, Cuellar WJ, Fuentes S, Kreuze JF, Gibson RW, Mukasa SB, Tugume AK, Tairo FD, Valkonen JPT (2012). Sweet potato Viruses: 15 Years of Progress on Understanding and Managing Complex Diseases. Plant Dis. 96(2):168-185.
Crossref
|
|
|
|
|
Clarke JD (2009). Cetyltrimethyl ammonium bromide (CTAB) DNA miniprep for plant DNA isolation. Cold Spring Harb Protoc. 2009(3):pdb-prot5177.
|
|
|
|
|
Harrison B, Robinson D (1999). Natural genomic and antigenic variation in whitefly-transmitted Geminiviruses (Begomoviruses). Annu. Rev. Phytopathol. 37(1):369-398.
Crossref
|
|
|
|
|
Kays S, Wang Y, McLaurin W (2005). Chemical and geographical assessment of the sweetness of the cultivated sweet potato clones of the world. J. Am. Soc. Hortic. Sci. 130(4):591-597.
|
|
|
|
|
Li R, Salih S, Hurtt S (2004). Detection of geminiviru.ses in sweet potato by polymerase chain reaction. Plant Dis. 88(12):1347-1351.
Crossref
|
|
|
|
|
Lotrakul P, Valverde R, Clark C, Sim J, De La Torre R (1998). Detection of a geminivirus infecting sweet potato in the United States. Plant Dis. 82(11):1253-1257.
Crossref
|
|
|
|
|
Luan Y, Zhang J, Liu D, Li W (2007). Molecular characterization of Sweet potato leaf curl virus isolate from China (SPLCV-CN) and its phylogenetic relationship with other members of the Geminiviridae. Virus Genes 35(2):379-385.
Crossref
|
|
|
|
|
Maina S, Edwards, OR, Barbetti, MJ, de Almeida, L, Ximenes, A, Jones RAC (2016). Deep sequencing reveals complete genome of Sweet potato virus G from East Timor. Genome Announc. 4 (5):e00957-16.
|
|
|
|
|
Mbanzibwa D, Tugume A, Chiunga E, Mark D, Tairo F (2014). Small RNA deep sequencing-based detection and further evidence of DNA viruses infecting sweet potato plants in Tanzania. Ann. Appl. Biol. 165(3):329-339.
Crossref
|
|
|
|
|
Miano D, LaBonte D, Clark C, Valverde R, Hoy M, Hurtt S, Li R (2006). First Report of a Begomovirus Infecting sweet potato in Kenya. Plant Dis. 90(6):832-832.
Crossref
|
|
|
|
|
Njeru R, Mburu M, Cheramgoi E, Gibson R, Kiburi Z, Obudho E, Yobera D (2004). Studies on the physiological effects of viruses on sweet potato yield in Kenya. Ann. Appl. Biol. 145(1):71-76.
Crossref
|
|
|
|
|
O'Brien P (1972). The sweet Potato: Its Origin and Dispersal. Am. Anthropol. 74 (3):342-365.
Crossref
|
|
|
|
|
Okada Y, Kobayashi A, Tabuchi H, Kuranouchi T (2017). Review of major sweet potato pests in Japan, with information on resistance breeding programs. Breed. Sci. 67(1):73-82.
Crossref
|
|
|
|
|
Padidam M, Sawyer S, Fauquet C (1999). Possible emergence of new geminiviruses by frequent recombination. Virology 265(2):218-225.
Crossref
|
|
|
|
|
Saitou N, Nei M (1987). The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4(4):406-425.
|
|
|
|
|
Tamura K, Nei M (1993) Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol. Biol. Evol. 10(3):512-526.
|
|
|
|
|
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013). MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol. Biol. Evol. 30(12):2725-2729.
Crossref
|
|
|
|
|
Thornton P, Jones P, Alagarswamy G, Andresen J (2009). Spatial variation of crop yield response to climate change in East Africa. Glob. Environ. Change 19(1):54-65.
Crossref
|
|
|
|
|
Thresh J, Fargette D, Jeger M (2003). Epidemiology of tropical plant viruses. In. Loebenstein G, Thottappilly G (eds) Virus and virus-like diseases of major crops in developing countries. Dordrecht: Kluwer Academic Publishers. Pp. 55-77.
Crossref
|
|
|
|
|
Valverde R, Clark C, Fauquet C (2003). Properties of a Begomoviruses Isolated from Sweet Potato [Ipomoea batatas (L.) Lam.] infected with Sweet potato leaf curl virus. Rev. Mex. Fitopatol. 21:128-136.
|
|
|
|
|
Valverde R, Clark C, Valkonen J (2007). Viruses and virus disease complexes of sweet potato. Plant Viruses 1(1):116-126.
|
|
|
|
|
Wasswa P, Otto B, Maruthi M, Mukasa S, Monger W, Gibson R (2011). First identification of a sweet potato begomovirus (sweepovirus) in Uganda: characterization, detection and distribution. Plant Pathol. 60(6):1030-1039.
Crossref
|
|
|
|
|
Zuckerkandl E, Pauling L (1965). Molecules as documents of evolutionary history. J. Theor. Biol. 8(2):357-366.
Crossref
|
|