ABSTRACT
The African forest, which was and still is the heart of biodiversity, is fast losing its bio-components due to natural disasters and human encroachment. A catalogue of the bio-components of the African forest is imminent so that conservation programs can be established to safeguard rare germplasm(s) from extinction. The population density of wild Auricularia (Mushroom) of Southwestern Nigeria was the focus of this research. DNA primers were obtained from Operon Technology, Alameda, California, USA. Characterization was done using phenotype, PCR and electrophoresis gel (RAPD). OPB-11 to OPB-21, OPH-3 to OPH-15, and OPT-1 to OPT-19 primers formed polymorphic bands with DNA samples of the specimens. Three (3) distinct species of Auricularia were identified in the forest of Southwest Nigeria based on PCR and RAPD analyses. Geospatial analysis showed that Auricularia auricula was present in the forests of Ekiti, Osun, Ogun, Oyo and Lagos states, but none was identified in Ondo State. On the contrary, Auricularia polytricha was only identified in the tropical rainforest and grassland vegetation of Ondo, Lagos and Oyo states.
Key words: Auricularia, mushrooms, population density, Southwestern Nigeria, phenotypic description, genome similarity index.
The oldest documentary of mushroom scavenging by humans was discovered in Tassili (Southern Algeria), Acacus (Libya) and Ennedi (Chad), etc., where the historical data was well preserved in magnificent rock drawings, paintings and other artifacts of the earliest inhabitants of Africa (also known as the cradle of humanity). So far, the mycoflora of modern Africa is still largely unknown (Gram, 2021). Mushroom foraging was and still remain a promising means of generating income within rural settlements in Africa, since it is a highly-sought-after nutritional (food) supplement, and a pertinent raw material for folk medicine (Degreef et al., 2016). In ancient times, the African woodland terrain was teeming with different variety of micro- and macroflora capable of biodegradation, such that the problem of waste management never arose since these “natural recyclers” efficiently and effortlessly transformed the huge amount of wastes deposited indiscriminately, on daily basis, in the environment (Degreef et al., 2016; Etaware, 2021). These florae (basically macro-fungi) were capable of degrading recalcitrant wastes and they even had the propensity to absorb and store toxic elements from the environment (Adenipekun et al., 2015; Etaware, 2021).
The macro-fungi (mushroom) population of the African forest is fast declining and they may become endangered in the nearest future due to extermination of their physical domain by climate change, natural disaster, animal and human intervention (Osemwegie et al., 2014). A direct catalogue and geo-referencing of the existing mushroom species within the African forest is imminent so that anti-grazing or anti-poaching laws refraining human activities in the coveted areas can be promulgated and other conservation programs can be established to safeguard the existing mushroom germplasms and prevent them from going extinct. Auricualaria species are highly priced mushrooms with nutritional and medicinal benefits (Chang and Hayes, 2013), they are currently ranked third in global mushroom production behind Lentinula and Pleurotus species (Royse et al., 2017; Bandara et al., 2019). In terms of distribution, Auricularia spp. are cosmopolitan, sadly, the species diversity within the genus Auricularia is on the decline. Currently, there are only less than 10 identifiable Auricularia spp. in China and a little below 25 species existing around the world (Wu et al., 2015).
In the rural and sub-urban areas of Africa, the most reliable means of identification of edible mushrooms by the indigens are mostly through phenotypic (physical) appearance, based on innate experience acquired through long term mushroom foraging in the wild. This is very risky, as the difference between a poisonous and edible mushroom is not always defined by physical appearance. Also, some researchers have been able to classify mushrooms based on physical appearance alone (Onyango et al., 2011). The use of physical appearance in the identification of edible mushrooms (Auricularia spp. inclusive) in some parts of Africa (especially Nigeria) is quite risky, unscientific, unreliable, misleading and may be ineffective on the long-run, since phenotypic classifications can be greatly influenced or affected by slight variation of environmental factors. Therefore, the current research was setup to identify the existing population of Auricularia spp. within the tropical forest of Southwestern Nigeria using sophisticated molecular techniques (PCR and RAPD), with a view to ensure proper documentation of the available species of Auricularia in the wild and their geographical location. Part of the aim of this research was to ensure that the mushroom germplasms collected for conservation were properly identified (Moore, 2013).
Taxonomy of Auricularia
Kingdom: Fungi
Division: Eumycota (Basidiomycota)
Sub-division: Basidiomycotina
Class: Agaricomycetes
Order: Auriculariales
Family: Auriculariaceae
Genus: Auricularia
Research area
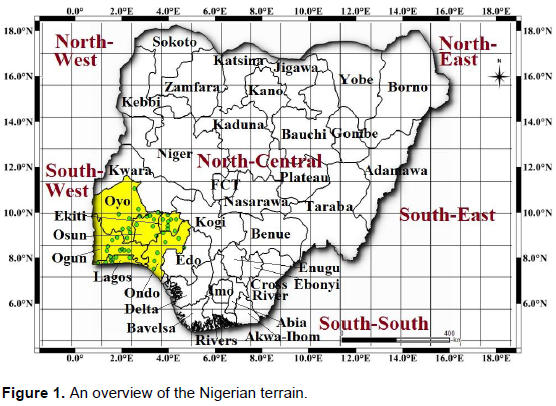
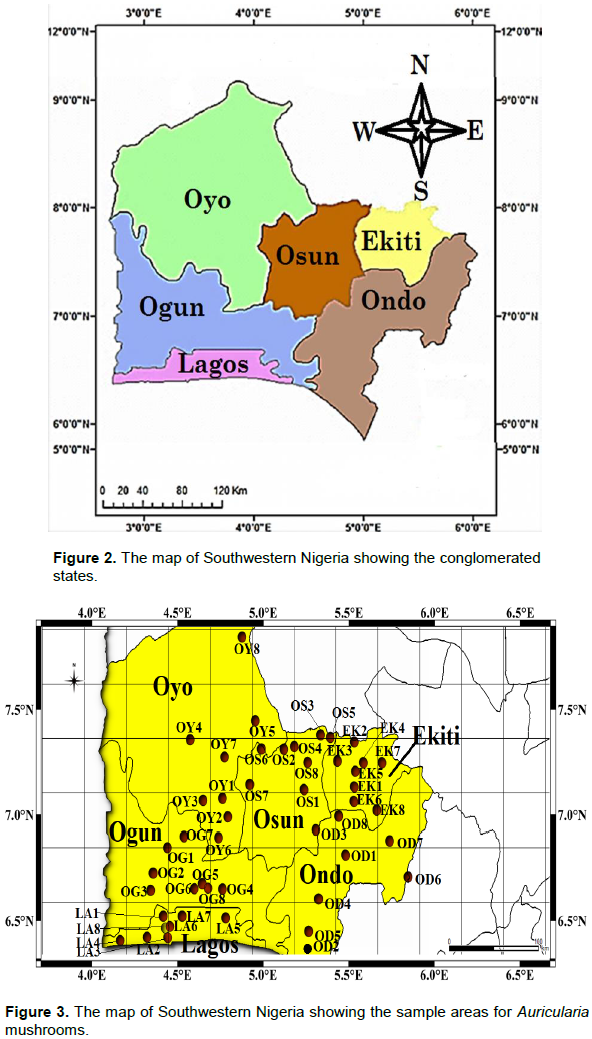
The tropical rainforest, mangrove swamp and savannah vegetation of the Southwestern region of Nigeria (Figure 1) was selected as the case study of this research. The zone is characterized by a bi-seasonal weather condition (Rainy and dry season), with sufficient annual precipitation (rainfall), ambient sunshine, high relative humidity and a warm day time temperature. Southwest Nigeria is one of the six (6) geo-political zones of the country, located on Latitude 9.081999° N and Longitude 8.675277°E, respectively. It is made up of six (6) states, that is, Ekiti, Lagos, Ogun, Ondo, Osun and Oyo states as shown in Figure 2.
The proposed sites for Auricularia catalogue in Southwest Nigeria
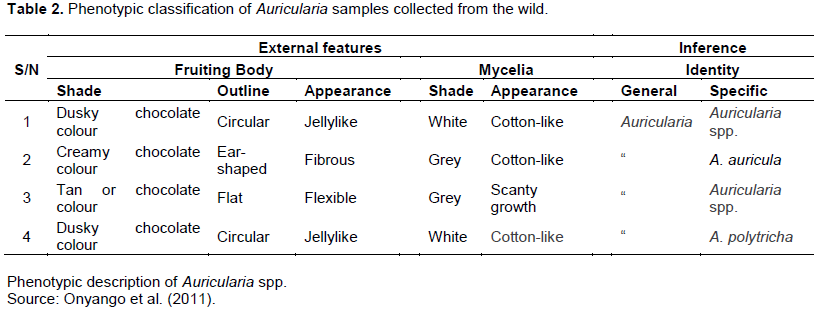
The proposed locations for mushroom foraging in Southwestern Nigeria were structured from the description given by renowned indigenous mushroom collectors. Sample stations were drawn from the six (6) states of Southwest Nigeria, covering all the possible terrain and forest locations within the earmarked area where mushrooms can thrive (Table 1). The forest and savannah (grassland) region of Southwest Nigeria was thoroughly surveyed towards the end of the rainy season in 2011 to the peak of the wet season in 2012. The features pertinent for effective identification of wild Auricularia spp. were adapted from the research of Musngi et al. (2004). The pattern of investigation of the wild forest of Southwest Nigeria is as shown in Figures 1 to 3.
Phenotypic identification of wild Auricularia
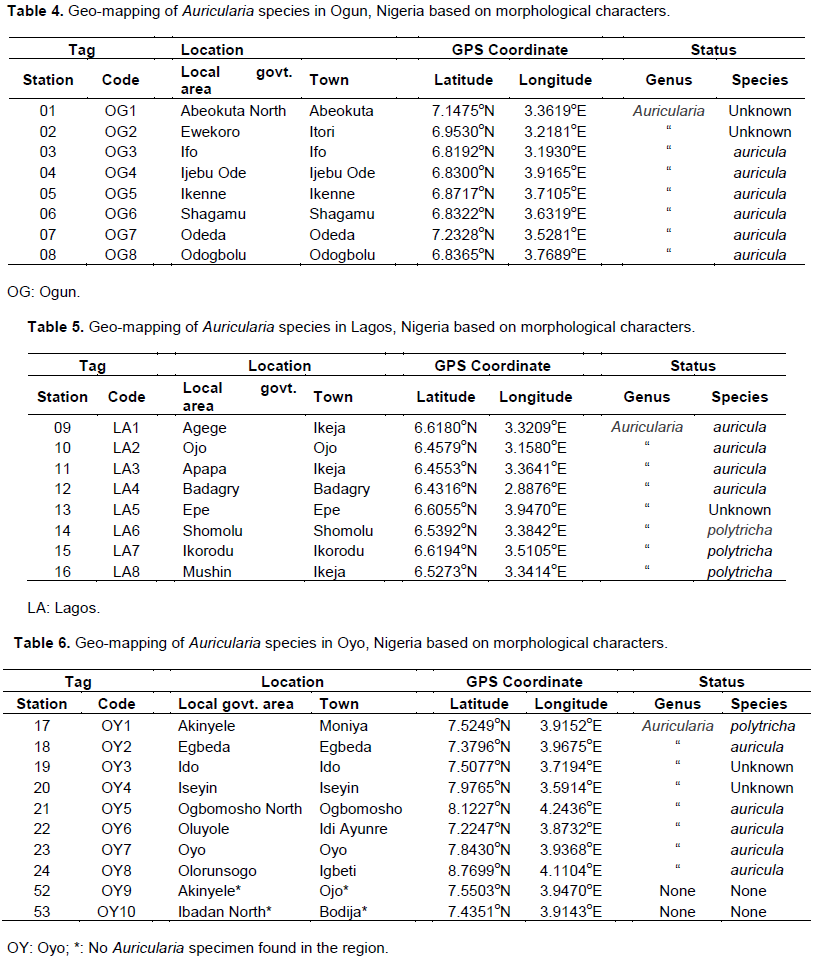
The identification of Auricularia spp. was done using both the traditional method (phenotypic characters only) and molecular characterization (using genome similarity). The traditional method was described in this session. A quick in-field identification was done by the researchers/collectors using the phenological description given by Onyango et al. (2011). The pre-identified Auricularia samples collected from the earmarked location within the wild forest of the six (6) states of Southwest geopolitical zone of Nigeria were aseptically packaged in sterile propylene bags and labelled appropriately (that is, sample site, coordinates, time of collection, and date). The collected samples were thereafter transferred to the mushroom house of the Department of Botany, University of Ibadan, Ibadan, Oyo State for further identification. Mushroom samples were sorted initially based phenotypic (morphological) characters with special focus on the appearance of the fruiting body and the presence or absence of reproductive structures (hymenium). The method adapted was a simplistic protocol used by native Africans in the identification of edible mushrooms (Auricularia spp. Inclusive), which was described by Onyango et al. (2011). The phenotypic characters investigated are shown in Table 2. Furthermore, anatomical studies of the collected samples were carried out in the laboratory, both at the University of Ibadan and National Institute of Horticulture (NIHORT), Ibadan, Oyo State, Nigeria. The spores from the basidiocarps of each mushroom was inoculated into full-strength PDA (in replicates of 3) and cultured for 7 days at 25±2°C, the mycelial morphology of the spawn produced were noted too.


Genome classification of the collected mushroom samples
Extraction and purification of mushroom DNA
Standard protocol for extraction of DNA from mushroom samples described by Chen et al. (2010) was adopted for this research. The steps involved are as follows:
(1) Aseptically excise about 200 mg of rehydrated tissue from the pileus of each mushroom sample
(2) Place the fragments of the excised tissue in an electric grinder with about 800 mL of CTAB buffer and pulverize to form a paste
(3) Incubate the pulverized samples at 65°C for 15 min in a water bath with constant agitation to allow even mixture
(4) Allow the mixture to cool before adding a composite solution of phenol, chloroform and iso-amyl in a ratio of 25:24:1
(5) Centrifuge the resulting mixture at 12,000 rpm for 15 min using a standard laboratory centrifuge
(6) Aseptically dispense the supernatant into sterile test tubes before adding about 400 μL of isopropanol (stored at 4°C).
(7) Homogenize the mixture by flipping manually up to five times to aid effective precipitation of the DNA
(8) Store at -80°C for 1 h.
(9) Centrifuge again at 12,000 rpm for 10 min to obtain DNA pellets.
(10) Decant the initial solvent from the pelleted DNA samples and rehydrate with 100 μL sterile distilled water and 2 μL of 10 mg/mL RNase
(11) Store at 4°C for 30 min (at this point, fragments of RNA strands will be dissolved completely).
DNA/RNA primer procurement
The primers used for this research were obtained from Operon Technology (Alameda, CA, USA) (Table 3).
Auricularia spp. genome profiling using PCR and RAPD
The genome identity of the pre-identified Auricularia samples collected in the wild was determined using PCR and profiled on electrophoresis gel (RAPD). The steps involved were described as the following.
PCR analysis
Reagents: A = Buffer (10x), B = 50 mM MgCl2, C = 2.5 mM dNTPs,
D = 500 U Taq DNA polymerase, E = Dimethyl sulfoxide (DMSO), and F = DEPC-treated water.
Procedure:
(1) An aliquot of 2.0 μL of the standardized DNA fragments (100 ng) was loaded into the tube of an Applied Bio-system thermo-cycler
(2) A mixture of 25 μL of A, 1.25 μL of B, 2.0 μL of C, and 0.2 μL of D were added to the loaded DNA fragment in the tube
(3) Furthermore, 1.0 μL of E, 1.0 μL of each primer and 16.05 μL of F were added to the mixture in the tube too.
(4) PCR amplification were conducted at 40-60 cycles at 94oC for 2 mins (initial cycling speed), 40 cycles at 94oC for 20 s, then at 72oC for 1 min., and at 54oC for 2 mins. Finally, run the sample at a speed of 40 cycles for 5mins at 72oC RAPD analysis
(5) An aliquot of about 2.5 μL of the already extracted and purified DNA sample obtained from the pre-identified Auricularia specimen was aseptically loaded into the compartments or wells of the RAPD device containing 1.5% agarose gel, for electrophoresis gel analysis.
(6) The loaded sample was observed under ultraviolet radiation (UV) to ascertain the quality of the samples extracted.
(7) The DNA fragments were separated based on their electron affinity and size
(8) Afterwards, sample uniformity was ensured by calibrating the separated DNA fragments to a value of 100 ng/μL.
Quantification of the calibrated and purified DNA samples
The Nano-Drop spectrophotometer (recommended) was used to quantify the already calibrated DNA samples obtained via the electrophoresis gel analysis. The criteria for proper quantification were stated thus:
(1) Sample size: 2 μL
(2) Optimum ratio (R): 1.8:2.0
(3) Optimum absorbance level/Optical density (OD): 260/280 nm
where Xcd = the intensity of the incident light and Ycd = the intensity of the transmitted light.
Statistical analysis
The data obtained from the electrophoresis gel analysis were scored as present (1) or absent (0). The dissimilarity matrix was ascertained by value of the Jaccard’s similarity coefficient (using the Jaccard Standard Protocol). Phylogenetic relations were determined by cluster analysis using UGPMA aided by the NTSYS-pc software version 2.02. Phylogenetic characterization into multivariate groups was done using principal component analysis (PCA) with Darwin software version 5.0.0.157 while polymorphic information content (PIC) was calculated using the method described by Botstein and colleagues in 1980.
Auricularia spp. stereotyping and mapping in Nigeria
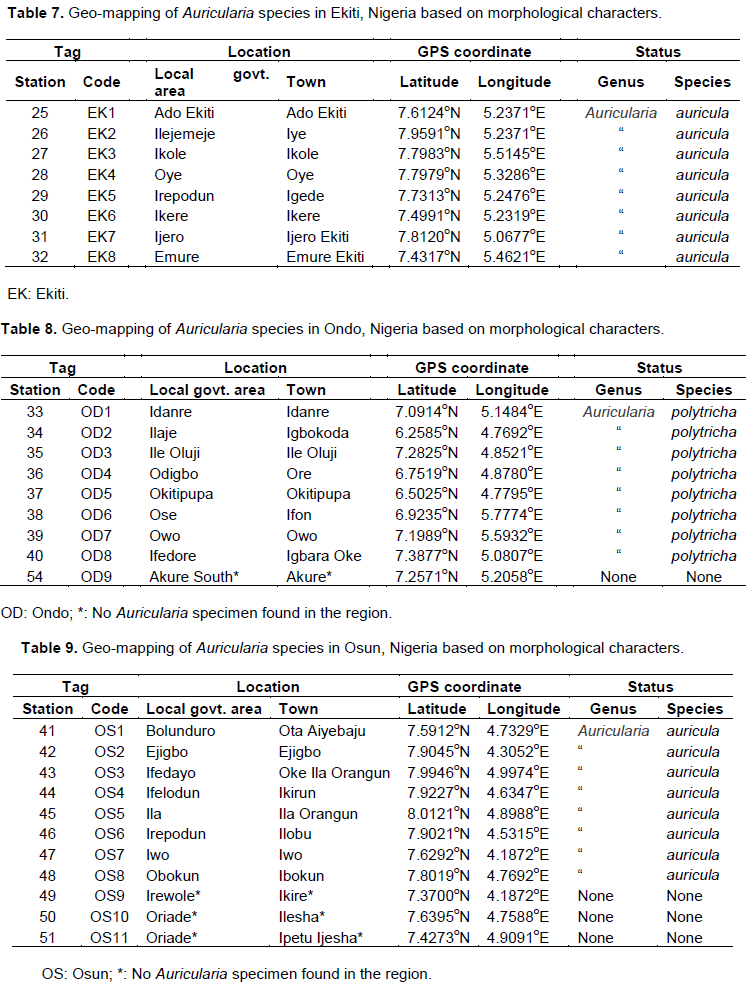
Auricularia spp. were identified in forty-eight (48) out of the fifty-four (54) locations initially earmarked for this research (Figure 3). A total of eight (8) locations were geo-referenced in each state where these edible mushrooms species can be found. It was noted that Auricularia spp. were abundantly present in the forest and grassland region of Abeokuta, Itori, Ifo, Ijebu Ode, Ikenne, Shagamu, Odeda, and Odogbolu in Ogun State (Table 4), Agege, Ojo, Apapa, Badagry, Epe, Shomolu, Ikorodu, and Mushin in Lagos State (Table 5), Moniya, Egbeda, Ido, Iseyin, Ogbomosho, Idi Ayunre, Oyo, and Igbeti in Oyo State (Table 6), Ado Ekiti, Iye, Ikole, Oye, Igede, Ikere, Ijero Ekiti, and Emure Ekiti in Ekiti State (Table 7), Idanre, Igbokoda, Ile Oluji, Ore, Okitipupa, Ifon, Owo, and Igbara Oke in Ondo State (Table 8), and finally, it was also sighted in places like Bolunduro, Ejigbo, Ifedayo, Ifelodu, Ila, Irepodun, Iwo, and Obokun in Osun State (Table 9).
The species A. auricula was identified in 6 communities in Ogun State (Ifo, Ijebu Ode, Ikenne, Shagamu, Odeda, and Odogbolu) (Table 4), 4 communities in Lagos (Agege, Ojo, Apapa, and Badagry) (Table 5), 5 communities in Oyo State (Egbeda, Ogbomosho, Idi Ayunre, Oyo, and Igbeti) (Table 6), 8 communities in Ekiti and Osun States (Ado Ekiti, Iye, Ikole, Oye, Igede, Ikere, Ijero Ekiti, Emure Ekiti) (Table 7), Bolunduro, Ejigbo, Ifedayo, Ifelodu, Ila, Irepodun, Iwo, and Obokun (Table 9), respectively. None was recorded for Ondo State (Table 8). On the contrary, A. polytricha was found in abundance in all the locations in Ondo State, that is, Idanre, Igbokoda, Ile Oluji, Ore, Okitipupa, Ifon, Owo, and Igbara Oke (Table 8), 3 locations in Lagos State (Shomolu, Ikorodu and Mushin) (Table 5), and one (1) location in Oyo State that is, Moniya (Table 6). The research team was unable to identify a rare Auricularia spp. in some part of the research area (Tables 4 to 6), further genomic analyses are required to effectively identify this rare species.
Genome classification of the collected mushroom samples
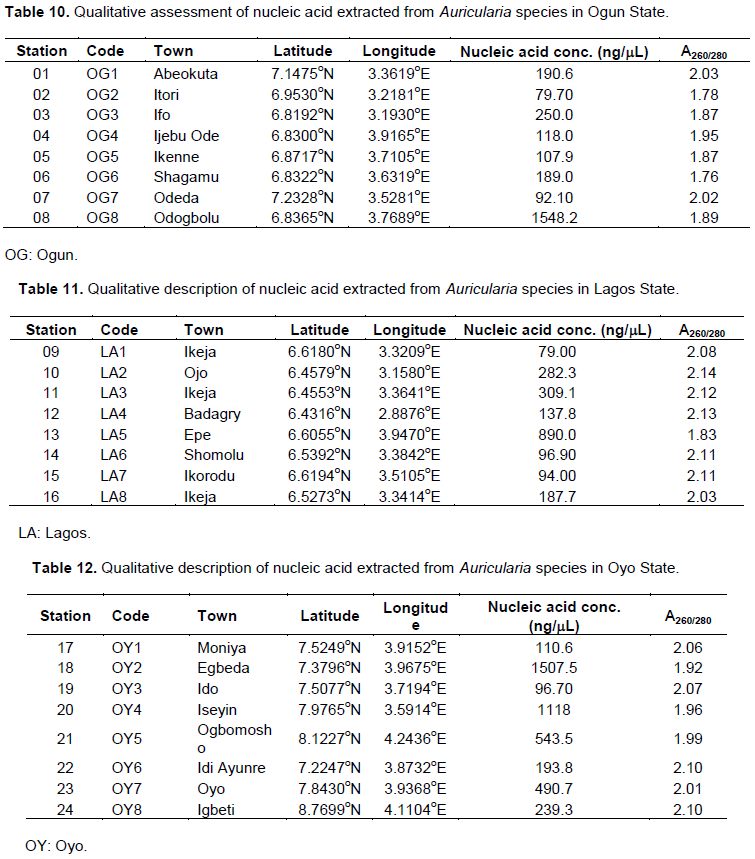
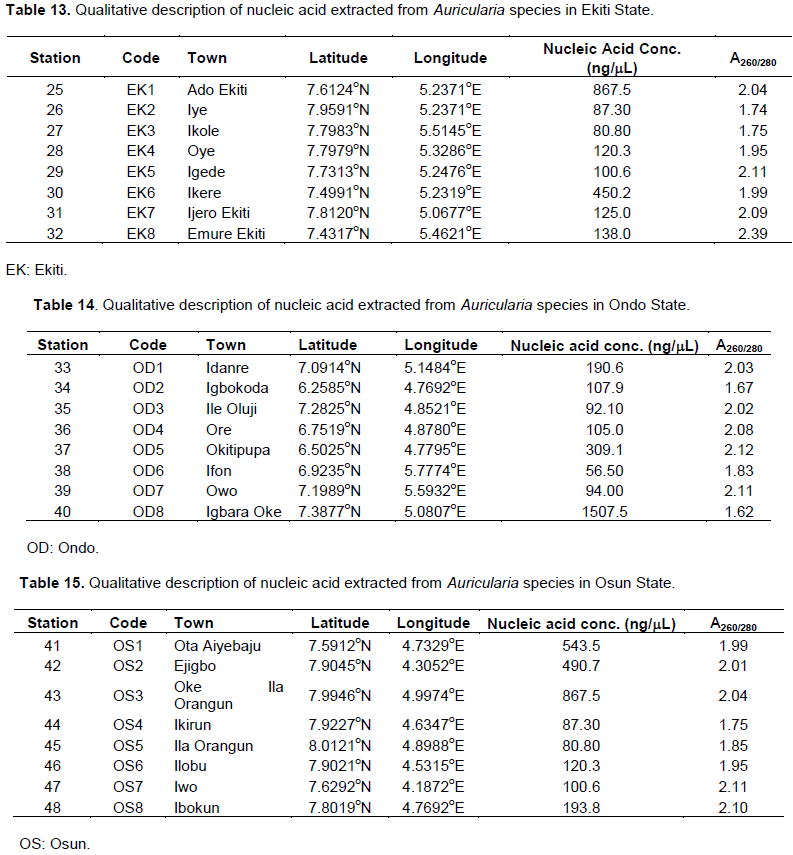
Quality assessment of the extracted Auricularia DNA
Majority of the DNA samples extracted for use in this experiment were pure (A260/280 ≥ 1.8), that is, high-quality DNA extracts, with the exception of OG2 (A260/280 = 1.78), OG6 (A260/280 = 1.76) (Table 10), EK2 (A260/280 = 1.74), EK3 (A260/280 = 1.75) (Table 13), OD2 (A260/280 = 1.67), OD8 (A260/280 = 1.62) (Table 14), and OS4 (A260/280 = 1.75) (Table 15), with little protein and RNA contaminants found in their DNA extracts. All the DNA samples extracted from the prospective Auricularia specimen collected from the tropical forest in Lagos and Oyo states were pure, with minimal impurities, that is, A260/280 ≥ 1.8 (Tables 11 and 12). It was observed that Auricularia specimen collected from Station 8 in Ogun State had the highest quantity of pelleted DNA sample with 1,548.2ng/mL of pure concentrated nucleic acid (Table 10). The least quantity of DNA extracts was obtained from Auricularia mushroom samples within Station 6 in Ondo State (56.5ng/mL of Nucleic acid concentration) (Table 14).
Genetic variability within the genus “Auricularia”
The major allele frquency of distribution of DNA fragments was highest on the primer location “OPH-15” (60%) and it was least on the primer location “OPB-15” (35%). The highest number of allele recorded during the genomic analysis was highest on the primer locations “OPB-21 and OPT-10” with a joint top score of 16. The least recorded number of allele was on primer loci “OPH- 10 and OPH-15” with 5 allele each. The highest level of genetic diversity observed during the genomic analysis was on primer locus “OPB-12” with a potential genetic variability of 80%, while the least was on the primer location “OPH-15” with an average genetic variability of 59%. The highest level of DNA band polymorphism was observed on primer locus “OPB-12” with 78% propensity for DNA band polymorphism. The least polymorphic information was noted at primer locus “OPH-15” with 56% chances of DNA polymorphism. Finally, the highest polymorphic amplicon was observed on primer locus “OPB-12” with an amplified value of 78.2%, while the least DNA polymorphic amplicon was observed on primer “OPH-15” with a value of 55.9% (Table 16).
Assessment of polymorphic nucleotide band formation
The profiled DNA amplicons showed little or no polymorphism at higher base pair units (Table 17). It was observed that none of the primers used in this research (except OPB-21 with 45/48 polymorphic DNA bands) had polymorphic bands at 900 bp. Subsequently, primers OPB-20 and OPB-21 were the only primers that registered polymorphism at 800 bp, that is, 46/48 and 9/48 polymorphic DNA bands, respectively. At 700 base pair units, five (5) out of the fourteen (14) primers investigated, were associated with polymorphic DNA activities, that is, OPB-11 (6/48 bands, as shown in Figure 4), OPB-12 (22/48 bands, as shown in Figure 5), OPB-20 (43/48 bands, as shown in Figure 7), OPB-21 (12/48 bands, as shown in Figure 8) and OPT-10 with 38/48 polymorphic DNA bands (Table 17). At lower base pair units, majority of the DNA primers were able to record DNA band polymorphism on electrophoresis gel.
At 600 bp, eight (8) primers recorded polymorphic activities on electrophoresis gel, that is, OPB-11 (13/48 bands), OPB-12 (2/48 bands), OPB-15 (2/48 bands), OPB-21 (37/48 bands), OPT-1 (9/48 bands), OPT-10 (41/48 bands) and OPT-19 (30/48 bands, as shown in Figure 10). At 500 bp, ten (10) DNA primers, including OPB-15 with 26/48 DNA bands (Figure 6), recorded polymorphic band formation, at 400 bp, thirteen (13) out of the fourteen (14) DNA primers recorded polymorphic activities, OPH-10 DNA primer showed the first sign of polymorphism at 400 bp with 3/48 DNA bands (Figure 9). Finally, at 300, 200, and 100 bp, all the primers used for this experiment showed significant levels of band formation when compared with the amplified DNA fragments from the wild Auricularia specimens (Table 17). The DNA primers OPB-20 and OPB-21 best described the nucleotide polymorphism with the investigated genome of the wild Auricularia specimens collected from the wild forests of Southwest, Nigeria.
The efficiency of the DNA primers used for this research
The DNA primers OPB-11, OPB-15, OPB-20, OPB-21, OPT-1 and OPT-5 were able to effectively detect 90 to 99% polymorphism in the DNA strands of the Auricularia specimen profiled on electrophoresis gel (Table 18), as such they were listed as the best markers for this research. Other DNA markers such as OPH-3, OPT-10 and OPT-19 showed an impressive 80 to 89% variability within the Auricularia population in Southwest Nigeria, while OPB-12 and OPH-5 were able to give between 70 and 79% variation in the examined Auricularia mushroom population. DNA markers OPH-10 and OPT-7 each gave between 60 and 69% variation, while OPH-15 was only able to detect between 10 and 19% variation at maximum in the Auricularia mushroom population of Southwest Nigeria (Table 18).
Grouping of Auricularia spp. in Nigeria based on genotype
The population of all the Auricularia mushrooms currently present in the six (6) states of Southwest Nigeria were effectively classified into six (6) clusters on the genetic dissimilarity chart (Figure 11) using PCR and RAPD markers on representative samples collected during field survey. The six (6) clusters of mushroom categories were effectively characterized into three (3) distinct species and further sub-classified into five (5) cultivars (sub- OS1, OS2, OS3, OS4, OS5, OS6, OS7, OS8, EK1, EK2, EK3, EK4, EK5, EK6, EK7, EK8 Cultivar II (Group V): OY2, LA1, LA2, LA3, LA4, OY5, OY6, OY7, OY8
Species 3: Unknown (Outliers)
OY3, OY4
The farther apart the Auricularia mushrooms on the genetic dissimilarity tree, the more related the species.
Thirty-one (31) locations were mapped in the wild where A. auricula can be found in the forest of Southwestern Nigeria. The geospatial analysis conducted showed that A. auricula was dominant in the forest of Ekiti state within the fringes of Ado Ekiti, Iye, Ikole, Oye, Igede, Ikere, Ijero Ekiti, and Emure Ekiti. Also, the same species of Auricularia was evenly distributed around the forest vegetation of Ota Aiyebaju, Ejigbo, Oke Ila Orangun, Ikirun, Ila Orangun, Ilobu, Iwo, and Ibokun (in Osun State), Ifo, Ijebu Ode, Ikenne, Shagamu, Odeda, and Odogbolu (in Ogun State), Egbeda, Ogbomosho, Idi Ayunre, Oyo, and Igbeti (in Oyo State) and Agege, Ojo, Apapa, Badagry, Epe, Shomolu, Ikorodu and Mushin (in Lagos State). There was no trace of A. auricula in the forest vicinity of Ondo State as at the time of filing this report. The unusual absence of this species of Auricularia in the rainforest and Savannah vegetations of Ondo State might be due to over exploration of this particular species for food and folklore medicine by the indigenous people of Ondo State, or maybe due to unfavourable weather conditions. In any case, the rationale behind the total absence of this species in Ondo State is still unclear and more scientific probing should be setup to explain this observation. The findings of this study were in concordance with the report of Adeniyi et al. (2018), who identified the carpophores of A. auricula-judae in ligneous habitat in Southwest Nigeria.
In contrast to the observation noted for A. auricula in Ondo State, A. polytricha was found in abundance in the forest terrain and savannah region of Ondo State, within the environs of Idanre, Igbokoda, Ile Iluji, Ore, Okitipupa, Ose, Owo, and Ifedore, respectively. It was sparsely distributed in the fringes of Lagos State, in places like Shomolu, Ikorodu and Ikeja only. Unfortunately, this species of Auricularia was neither found in the forest of Ogun, Ekiti, Osun, nor Oyo states. This strange observation is a puzzle that requires answer. The observation noted in the current study was in line with the research by Titilawo et al. (2019) who collected samples of A. polytrichia from the ligneous (woody) substrates within the forest of Southwestern Nigeria, in order to determine its nutritional (proximate) and phytochemical properties. Further probing of the already identified Auricularia spp. was carried out using genome similarity aided by PCR and electrophoresis gel techniques (RAPD). The analysis showed that there was slight variation in the genetic makeup of the Auricularia (mushrooms) found in Southwestern Nigeria. The group was further subdivided into five (5) cultivars (sub-species), based on the genome similarity index. The taxonomic groups unearthed by this research are still under review. The findings from this research were supported by the research work of Ekun et al. (2018), who earlier reported a similar observation of an unidentified Auricularia (mushroom) found deep in the tropical rainforest of Southwest Nigeria.
The study was able to establish the fact that the population of A. polytricha in the wild forests and grassland vegetations of Southwest Nigeria are on the decline and that efforts geared towards rescuing and conservation of the germplasm of that particular Auricularia spp. should be put in place soonest. Also, an interesting development unravelled by this research was the collection of rare species of Auricularia mushrooms; although, these rare species are few and sparsely distributed around Southwest Nigeria, it is also pertinent that they are protected and conserved too. Laws involving selective or restricted mushroom poaching or foraging in the rural areas of Nigeria should be promulgated so that some of these endangered mushroom species can be protected in their natural habitat.
The authors have not declared conflict of interests.
REFERENCES
|
Adenipekun CO, Ipeaiyeda AR, Olayonwa AJ, Egbewale SO (2015). Biodegradation of Polycyclic Aromatic hydrocarbons (PAHS) in spent and fresh cuttings fluids contaminated soils by Pleurotus pulmonarius (Fries).Quelet and Pleurotus ostreatus (Jacq) Fr.Kumm. African Journal of Biotechnology 14(8):661-667.
Crossref
|
|
|
|
Adeniyi M, Odeyemi Y, Odeyemi O (2018). Ecology, diversity and seasonal distribution of wild mushrooms in a Nigerian tropical forest reserve. Biodiversitas Journal of Biological Diversity 19 (1):285-295.
Crossref
|
|
|
|
|
Bandara AR, Rapior S, Mortimer PE, Kakumyan P, Hyde KO, Xu J (2019). A review of the polysaccharide, protein and selected nutrient content of Auricularia, and their potential pharmacological value. Mycosphere Journal 10(1):579-607.
Crossref
|
|
|
|
|
Chang ST, Hayes WA (2013). The biology and cultivation of edible mushrooms. Academic Press, London, UK 5 p.
|
|
|
|
|
Chen H, Rangasamy M, Tan SY, Wang H, Siegfried BD (2010). Evaluation of five methods for total DNA extraction from western corn rootworm beetles. PLoS one 5(8):e11963.
Crossref
|
|
|
|
|
Degreef J, Demuynck L, Mukandera A, Nyirandayambaje G, Nzigidahera B, De Kesel A (2016). Wild edible mushrooms, a valuable resource for food security and rural development in Burundi and Rwanda. Biotechnology, Agronomy, Society and Environment 20(4):441-452.
|
|
|
|
|
Ekun VS, Adenipekun CO, Ogunkanmi LA, Ojuederie OB, Igwe DO (2018). Molecular characterization of Auricularia spp from southwestern Nigeria using Random Amplified Polymorphic DNA (RAPD) markers. Nigerian Journal of Biotechnology 35(1):34-43.
Crossref
|
|
|
|
|
Etaware PM (2021). Biomineralization of toxicants, recalcitrant and radioactive wastes in the environment using genetically modified organisms. Journal of Agricultural Research Advances 3(2):28-36.
|
|
|
|
|
Gram H (2021). Intoxications and the oldest known mushroom cult in Africa. Accessed on August 26, 2021 at 17:16 GMT. 9pg.
View
|
|
|
|
|
Moore M (2013). Mushrooms: taste of the earth. Food and Nutrition 5 p.
|
|
|
|
|
Musngi RB, Lalap AL, Reyes RG (2004). Four species of wild Auricularia in Central Africa. Journal of Tropical Biology 3:49-51.
|
|
|
|
|
Onyango BO, Arama PF, Palapala VA, Wagai SO, Gichimu BM (2011). Suitability of selected supplemented substrates for cultivation of Kenyan native wood ear mushrooms (Auricularia auricula). American Journal of Food Technology 6(5):395-403.
Crossref
|
|
|
|
|
Osemwegie OO, Okhuoya AJ, Dania AT (2014). Ethnomycological Conspectus of West African Mushrooms: An Awareness Document. Advances in Microbiology 14(1):39-54.
Crossref
|
|
|
|
|
Royse DJ, Baars J, Tan Q (2017). Current overview of mushroom production in the world. In: Zied, D. C., Pardo-Gimenez, A. (eds). Edible and Medicinal mushrooms: technology and applications. Wiley, Hoboken.
Crossref
|
|
|
|
|
Titilawo MA, Oluduro AO, Odeyemi O (2019). Proximate and chemical properties of some underutilized Nigerian wild mushrooms. JMBFS Preprint. Accessed on September 03, 2021 at 18:25 GMT. 1-13.
View
|
|
|
|
|
Wu F, Yuan Y, He SH, Bandara AR, Hyde KD, Malysheva VF, Li DW, Dai YC (2015). Global diversity and taxonomy of the Auricularia auricula-judae complex (Auriculariales, Basidiomycota). Mycological Progress 14(95):1-16.
Crossref
|
|