ABSTRACT
Sorghum bicolor L. is an important crop in many tropical regions, yet it has received little attention in applying modern biotechnologies for improvement due to transformation difficulties. Drought is among the most important factors limiting sorghum productivity. The Nicotina Protein Kinase 1 (NPK1) gene confers tolerance to adverse environmental conditions. The present work was conducted to determine the amenability of sorghum genotypes to Agrobacterium-mediated transformation with the NPK1 gene and to characterize the role of that gene in S. bicolor. The NPK1 gene along with the bar gene was successfully introduced into two sorghum genotypes: Dorado and SPGM94021. The transformation frequencies were 1.49 and 1.79% for Dorado and SPGM94021, respectively. Stable integration of the transgenes in T0 and T1 plants was confirmed through polymerase chain reaction (PCR) and Southern blotting. The NPK1 gene expression was measured through real-time-PCR. T1 plants tested in vitro for tolerance to mannitol osmotic stress maintaining a higher growth rate and showed increased tolerance to stress conditions compared to the non-transgenic plants. The transgenic sorghum had a significantly higher kernel weight under drought stress conditions than the control plants. Accordingly, the NPK1 gene might induce a mechanism that protects sorghum plants against possible water-deficiency stress conditions.
Key words: Abiotic stress related genes, regulatory genes, signal transduction, transgenic sorghum, plant transformation, quantitative real time-polymerase chain reaction (qRT-PCR), drought stress tolerance.
Sorghum [Sorghum bicolor (L.) Moench], a tropical plant of the family Poaceae, is the fifth most important cereal after wheat, rice, maize and barley and plays a unique role in food security and renewable energy (Belton and Taylor, 2004). The crop is well adapted to tropical and subtropical areas throughout the world. Beside its principal uses as flour, in the preparation of porridge and unleavened bread, sorghum species are sources of fiber and fuel and are, also, used in the alcohol industry as they are rich in starch. Despite its importance, the yield and quality of sorghum are constrained by a range of biotic and abiotic factors. Nevertheless, abiotic stresses remain the greatest constraints of crop production with approximately 70% estimated yield reduction (Acquaah, 2007). Drought is one of the priciest factors affecting a huge number of people every year (Wilhite, 2000). Drylands occupy about 41% of the global terrestrial surface and are home to more than a third of the world's population (Mortimore, 2009). Moreover, the increase of potential evapotranspiration and the changes of the precipitation patterns under a warming climate are leading to a growing global aridity and expansion of drylands (Milesi et al., 2010).
Therefore, interest in crop tolerance to environmental stresses has been growing since the last few decades (Tuberosa and Salvi, 2006). Sorghum is a drought tolerant crop and often grows where water stress conditions are expected. However, the yields, under dryland conditions, are severely affected and much less than that of irrigated sorghum (Assefa and Staggenborg, 2010). Several investigations have been conducted on grain sorghum under stress conditions. Seed mass, harvest index, and biomass were among the parameters affected, most severely, by moisture stress (Wenzel, 1999). Genetic engineering is a potential tool for crop improvement. Although grain sorghum is a very important cereal, particularly in the semi-arid tropic areas as a vital source of food for millions of people around the world, sorghum improvement through genetic engineering is progressing slowly. Like other cereal crops, the first fertile transgenic sorghum plants were obtained via particle bombardment (Casas et al., 1993). Agrobacterium-mediated transformation of sorghum was first reported seven years later (Zhao et al., 2000).
To improve sorghum abiotic stress tolerance, HVA1, thaumatin-like protein (TLP), mtld, OsCDPK-7 and TPS1 genes were successfully introduced into sorghum genome (Devi et al., 2004; Gao et al., 2005a; Maheswari et al., 2010; Mall et al., 2011; Yellisetty et al., 2015). In order to make plants more tolerant to stress and restore the cellular balance, transferring a single gene encoding a single specific stress protein, that is, "single-action" genes may not suffice the required tolerance levels. To overcome such constraint, plant transformation with regulatory genes has emerged (Bhatnagar-Mathur et al., 2008). Nicotina Protein Kinase 1 (NPK1) is a regulatory single transduction gene located upstream of the oxidative pathway and can induce expression of HSPs and GSTs (Kovtun et al., 2000; Shou et al., 2004a). Under stress conditions, plants generate reactive oxygen species, including hydrogen peroxides, superoxide anion, and hydroxyl radical (Shou et al., 2004a).
Hydrogen peroxide accumulation can initiate the expression of detoxification and stress protection genes, e.g. heat shock proteins (HSPs), glutathione-S-transferases (GSTs), peroxidases and pathogenesis-related proteins, thereby, guarding plants against stress damages (Kovtun et al., 2000). HSPs serve as molecular chaperones in the ATP-dependent protein assembly/disassembly and prevent protein denaturation during stress (Horwich, 2014). GSTs are enzymes that can detoxify endobiotic and xenobiotic compounds through covalent linkage of glutathione to hydrophobic substrates. Activation of these stress genes can reduce damage caused by chilling, heat, and drought; thus, protect plants from environmental stresses (Das and Roychoudhury, 2014; Li et al., 2003), thereby improving the yield potential of the major cereal crops. The objectives of this study were to utilize Agrobacterium-mediated transformation method to develop water use efficient sorghum lines and to evaluate the regenerated transgenic lines at molecular level. An additional goal was to evaluate the first generation of the transgenic lines (T1) for survival to water deficiency in vitro and under greenhouse conditions.
Plant materials, Agrobacterium tumefaciens strain and binary vector
Two S. bicolor genotypes, that is, Dorado and SPGM94021, obtained from the Department of Sorghum, Field Crops Research Institute, Agricultural Research Center, Egypt, were sown in the field during the sorghum season and plants have been self-pollinated. Panicles were harvested 10 to 12 days post pollination. A. tumefaciens strain LBA4404 containing the standard binary vector pSHX004 (Shou et al., 2004a) was used for sorghum transformation. The binary vector has been introduced into the A. tumefaciens strain LBA4404 by direct transformation of the competent cells with the pSHX004 DNA. The vector system, pSHX004 in LBA4404, was maintained on a yeast extract peptone (YEP) medium containing 50 mg/L spectinomycin and 25 mg/L rifampicin. Bacteria cultures, for weekly experiments, were initiated from stock plates stored for up to two weeks at 4°C.
Sorghum transformation
Sorghum panicles were surface sterilized as described in Assem et al. (2014). Immature embryos ranging in length from 1.0 to 1.5 mm were aseptically excised from kernels and prepared for transformation. The A. tumefaciens strain LBA4404 was used for the delivery of the binary vector pSHX004 into the dissected immature embryos. For transformation, A. tumefaciens cultures were grown for 2 days at 28°C on YEP medium amended with 100 mg/L spectinomycin and 25 mg/L rifampicin. One loop of the culture was scrapped and suspended in 5 ml of liquid infection medium supplemented with 100 μM acetosyringone. Immature zygotic embryos were washed twice with bacteria-free infection medium, the final wash was discarded and 1 to 1.5 ml of A. tumefaciens suspension (OD550 = 0.4-0.5) was added to the embryos.
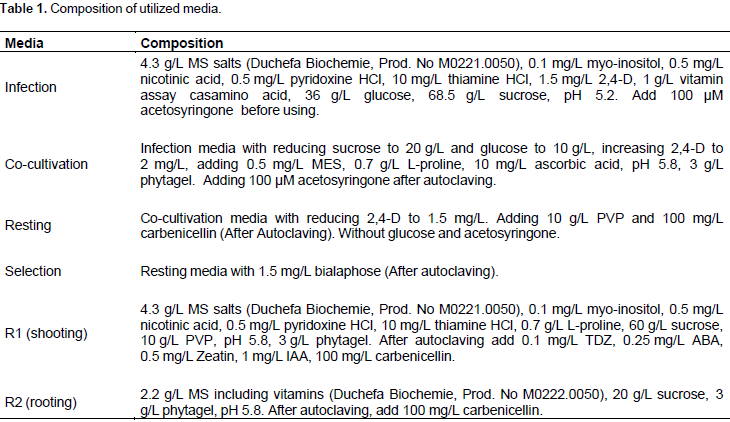
The tubes were incubated, in dark, for 10 min at ambient temperature. After infection, embryos were transferred to co-cultivation medium with scutellum side up. Plates were incubated in the dark at 22°C for 4 days after which embryos were transferred to resting medium and incubated at 28°C for one week. The transformation process can be divided into 5 sequential steps: agro-infection, co-cultivation, resting, selection, and plant regeneration. The infection and the co-cultivation media were prepared as described by Zhao et al. (2000), without modification. The compositions of all media utilized in this study are shown in Table 1.
Selection and regeneration
For stable transformation, selection of transformed events was carried out using selection medium containing 1.5 mg/L bialaphos as a selective agent. After 8 weeks on selection medium, survived embryogenic calli were transferred to regeneration medium (R1) for shoot development. Calli-derived shoots were transferred to rooting medium (R2). Cultures of shooting and rooting stages were maintained at 28°C under 16 h photoperiod cool white fluorescent light (75 µmol/m2/s). Regenerated plantlets have been transferred to soil for further development in the greenhouse. Putative transgenic and non-transgenic plants were subjected to leaf painting when 6 to 7 leaves had fully emerged from the whorl. Solution of the commercial Basta herbicide at concentration of 0.1% was applied to the upper and lower surfaces of the fifth fully emerged leaf.
Polymerase chain reaction (PCR)
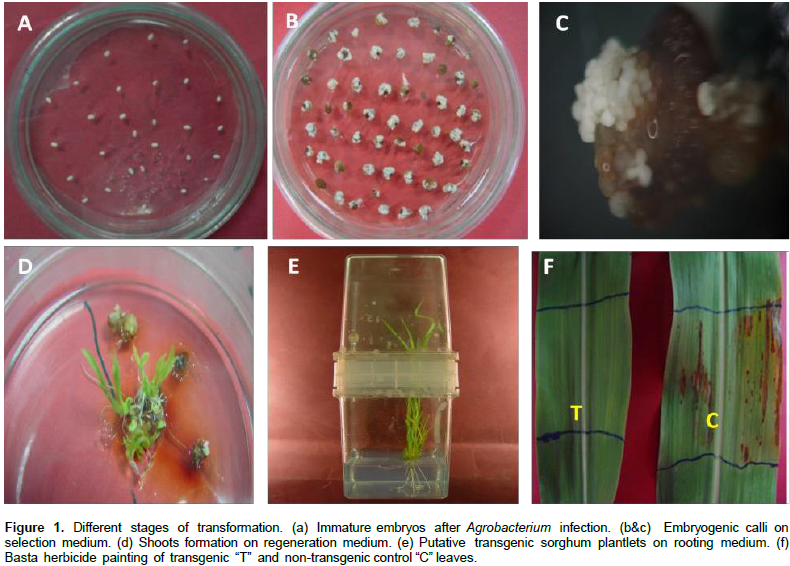
PCR analysis was conducted to screen the putatively transgenic T0 and T1 plants. Genomic DNA was extracted from putatively transgenic sorghum plants and non-transformed plants, as control, using the cetyltrimethylammonium bromide (CTAB) protocol (Murray and Thompson, 1980). The forward and reverse primers, 5' TAACAAATGGATGCTGAAGC 3' and 5' CCATCCCAACATAGTGAGAT 3', were used to amplify a 605 bp fragment of DNA containing part of the NPK1 transgene; whereas the forward and reverse primers, 5' TAC ATCGAGACAAGCACGGTCAACT 3' and 5' ACGTCATGCCAGT CCCGTG 3', were used to amplify a fragment of 484 bp for the bar gene. PCR reactions were carried out in a total volume of 25 μl containing 10 ng of genomic DNA, 1x PCR buffer, 3 mM MgCl2, 0.2 μM of forward and reverse primers, 0.24 mM dNTP, and 1.5 U of Taq polymerase. Amplification was conducted in a T100 thermocycler (BIO-RAD, Singapore) using the following program for the NPK1 fragment: An initial DNA denaturation for 5 min at 94°C followed by 35 amplification cycles (94°C, 1 min; 55°C, 1 min, 72°C, 1 min) and a final extension step at 72°C for 10 min. For the amplification of bar fragment, the annealing temperature was increased to 60°C for 1 min.
Southern blotting
Ten micrograms of genomic DNA from sorghum plants were used for Southern blot analysis. DNA from T0 and T1 Agrobacterium-derived events and control plants were digested with the restriction enzyme EcoRI, at 37°C overnight. Digested DNA was separated on a 0.8% (w:v) agarose gel and transferred by alkaline transfer method onto a positively charged nylon membrane (Boehringer, Mannheim, Germany) and then cross-linked to the membrane by UV irradiation. A fragment (605 bp) of the NPK1 gene sequence, generated by PCR, was DIG-labeled and used as a probe. The membrane was hybridized overnight at 68°C with the labeled probe. The hybridization signals were detected by the colorimetric method. The labeling and signal detection were performed using the random priming DNA labeling and detection kit (Roche Cat. No.11093657910).
Quantitative Real-Time PCR
Total RNA was extracted using RNeasy Plant Mini Kit (Qiagen, Hilden, Germany) following manufacturer’s instructions. After removing DNA using TURBO DNAse following the manufacturer’s instructions (Life Technologies), total RNA samples were quantified using the Qubit RNA assay (Life Technologies). One step-Taqman assay was used to detect the amount of NPK1 mRNA in the samples. The 18S small-subunit ribosomal RNA gene was used as the endogenous control for RNA quantification. NPK1 primers and probe were synthesized and labeled by Life Technologies (Carlsbad, CA). Real-time PCR NPK1 primers and probe sequences were obtained from Shou et al. (2004b) and 18S rRNA TaqMan assay mix was obtained from Life Technologies. Real-time PCR was carried out using the 7500 Fast Real-Time PCR instrument (Life Technologies).
Reactions were conducted in a total volume of 20 µl containing 100 ng of sorghum total RNA, 10 µl of 2X master mix from the TaqMan RNA-to-CT 1-step Kit (Life Technologies), 0.4 mM of each primer, 0.2 mM of probe, and 0.5 µl of reverse transcriptase from the same kit. PCR conditions were as follows: Reverse transcription on 48°C for 15 min, heat activation at 95°C for 10 min, followed by 40 amplification cycles (at 94°C for 30 s, annealing at 57°C for 1 min and extension at 72°C for 1 min). All samples were tested in triplicate. To eliminate the possibility of inherent variation in the amount of starting material between samples, the amounts of NPK1 mRNA detected by real-time RT-PCR were normalized by dividing them with their corresponding amounts of 18S rRNA.
Preliminary evaluation of transgenic plants under water-deficit conditions
The performance of the transgenic plants expressing the NPK1 gene was evaluated under mannitol stress conditions using seeds from the two transgenic lines Dorado and SPGM94021. Drought tolerance assay was applied as described in Kim et al. (2014) with modifications. Briefly, T1 seeds from each transgenic line and non-transgenic lines were surface-sterilized and germinated for four days in the dark at 25°C on half-strength MS basal medium containing 1% sucrose with or without 5 mg/L bialaphos for transgenic and non-transgenic plants, respectively. Germinated seedlings were then transferred to a half-strength MS containing 400 mM mannitol medium and maintained at 28°C under 16 h photoperiod cool white fluorescent light (75 µmol/m2/s). These experiments were replicated three times. The response of young seedlings to stress conditions was analyzed after 6 days.
The parameters of shoot length, root length and fresh weight were scored for twenty randomly selected bialaphos resistant seedlings and plantlets were transferred to a soil mixture composed of peatmoss: soil: sand (1:1:1) in pots of 10 cm diameters. Plantlets were grown in greenhouse at 28ËšC and 16/8 h (light/darkness) photoperiod. Plantlets were grown under well-watered conditions of 100% field capacity for the first three weeks. Soil water content was maintained by weighing and adding water into pots, daily. After three weeks of well irrigation, water withholding was applied for 11 to 13 days (depending on the weather) until leaves wilted, at which point the plants were irrigated. Repeated drought cycles were imposed until plants reached anthesis. The yield potential was evaluated by counting the number of kernels per panicle and weighing 100 grains harvested from the transgenic and control plants grown under drought stress conditions in the green house.
Statistical analysis
Three replicates of each experiment were analyzed for significant difference between treatments. For transformation experiments, to eliminate the variation in the initial number of transformed embryos, the percentage values of each replicate were compared in both genotypes. For the preliminary evaluation of T1 plants, the mean values of each parameter from twenty randomly selected plants for each replicate were compared relatively to the other genotype and to the nontransgenic control. Significance was determined by the analysis of variance (ANOVA) employing ASSISTAT software version 7.7 using a complete randomized design with repetition and the difference between the means were compared using Tukey test at a level of 5% probability.
Sorghum transformation and regeneration
Agrobacterium-mediated transformation was utilized to develop transgenic sorghum plants with low transgene copy number. Two sorghum genotypes; Dorado and B-SPGM49021 were transformed with the standard binary vector pSHX004 in LBA4404. The transgene of interest was the kinase domain of the NPK1 gene driven by a constitutive promoter (35S C4PPDK). The transformation data (Table 2) were obtained from three independent experiments for sorghum genotype Dorado and two independent experiments for genotype SPGM94021. Most of the infected immature embryos showed normal growth on co-cultivation and resting media, although some embryos failed to grow after transformation. Selection stage was initiated by placing the embryo-derived calli on selection media (callus induction media containing 1.5 mg/l bialaphos).
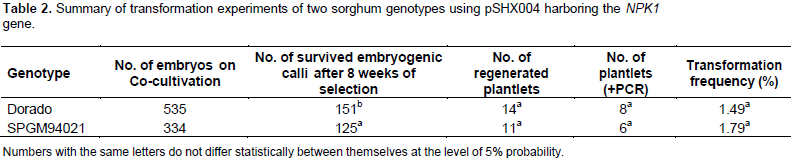
After two months of selection, survived embryogenic calli were used for shoot regeneration (Figure 1). Transgenic plants grew normally in the greenhouse like non-transgenic plants and showed normal fertility. The transformation frequencies based on PCR screening of the putatively transgenic plantlets were 1.79 and 1.49% for B-SPGM94021 and Dorado, respectively. Transformation frequency was calculated as the number of regenerated PCR-positive plantlets/number of agro-infected embryos × 100. Leaf painting assay indicated improved tolerance of the transgenic plants to Basta herbicide as compared to the non-transgenic plants which showed wilting at the painted area (Figure 1f). Transgenic T0 plants were harvested and T1 seeds of both genotypes were grown in the bio-containment greenhouse to test the inheritance of the transgenes.
Analysis of the transgenic plants
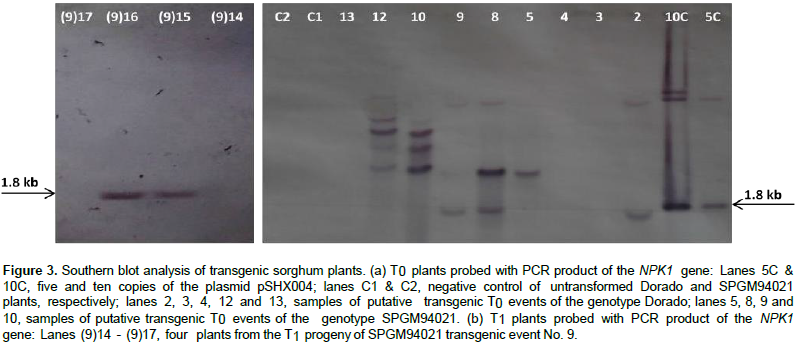
PCR analysis (Figure 2) revealed the presence of the expected bands for bar and NPK1 amplicons at 484 and 605 bp, respectively, in 14 out of a total of 25 individual events of the genotypes Dorado and SPGM94021 with no T-DNA truncation. Similarly, PCR analysis was conducted on the T1 plants. As shown in Figure 2c, bands corresponding to the expected size of 605 bp for the NPK1 gene were observed in some of the progeny, thus, confirming the inheritance of the transgene. To confirm the integration of the transgene, Southern blot analysis of T0 and T1 transgenic plants for NPK1 gene was conducted. The results confirmed the stable integration of the NPK1 gene into the sorghum genome as well as its transmission to the T1 plants (Figure 3).
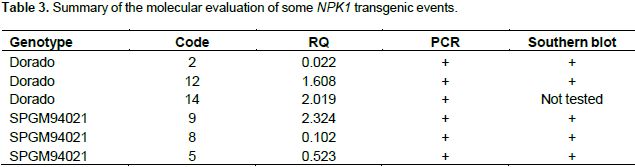
The estimation of transgene copy number revealed the presence of fewer than five copies of the NPK1 transgene, e.g. two copies in sample No. 2, four copies in sample No. 8, and one copy in sample No. 9. Further analysis of the putative transgenic plants using a quantitative real-time RT-PCR analysis indicated all the tested transgenic events expressed the NPK1 gene, although they had different expression levels. Table 3 summarizes the results of the molecular evaluation of 6 independent transgenic events, 3 belonging to the Dorado genetic background while the other 3 were generated from transforming the SPMG94021 background. All 6 events were positive in PCR evaluation and in all, except for event 14 (not tested), the band specific for the NPK1 transgene were detected in Southern blotting. Relative Quantification (RQ) represents the values of accumulated mRNA of the transgene compared to the endogenous gene of the calibrator sample (control non-transgenic) valued as 1.
Evaluation of T1 plants under drought stress conditions
In the present investigation, transgenic seeds were recovered from both sorghum cultivars, Dorado and SPGM94021. A preliminary kill curve experiment has been carried out with control seedlings of both genotypes on 200, 400 and 600 mM mannitol. Results revealed that 400 mM concentration of mannitol was very critical for control seedlings (Zamzam, 2014). Thus, a culture medium containing 400 mM mannitol was employed to evaluate the performance of T1 transgenic seedlings expressing NPK1 gene. As shown in Table 4 and Figure 4, transgenic plants maintained a relatively higher growth rate than that of the non-transgenic plants, under drought stress conditions. Significant differences in shoot length and fresh weight were found between non-transgenic and their counterparts’ transgenic plants expressing the NPK1 gene. However, the reduction in root lengths was not significant at 5% probability.
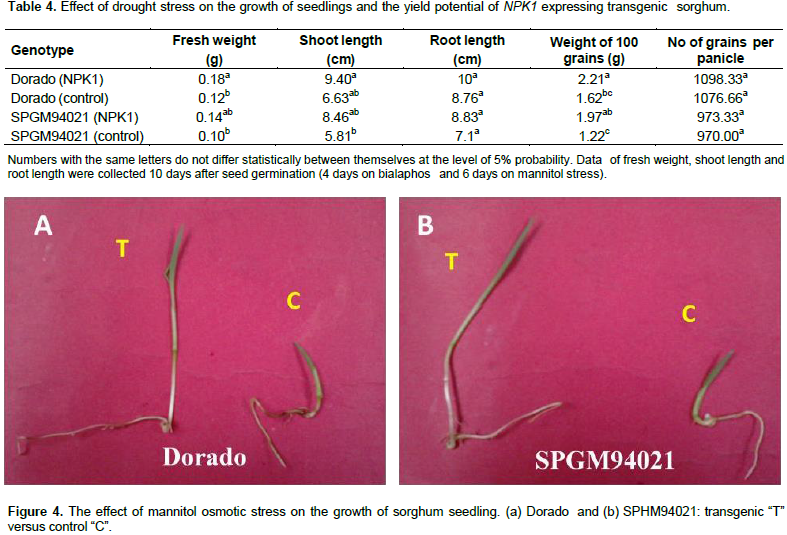
In the greenhouse, three-week-old control and transgenic plants were subjected to water withholding. Normally, under our greenhouse conditions, the non- transgenic sorghum plants are usually irrigated every 6 to 7 days. In this experiment, watering was stopped for 11 to 13 days. The transgenic plants displayed a higher capacity of recovering after re-irrigation compared to the non-transgenic plants. Moreover, the weight of 100 grains and the number of grains per panicle were used as physiological parameters for yield potential under water-deficit conditions. Kernels were collected from both transgenic plants and few recovered control plants after repeated exposure to drought stress under greenhouse conditions. The transgenic sorghum plants had a significantly higher mean of grain weight, under drought stress conditions, than the control plants.
Regulatory genes are very important research interest. Transcription factors represent an important category of regulatory genes. Many genes involved in stress response can be simultaneously regulated by a single gene encoding stress inducible transcription factor (Kasuga et al., 1999). While the expression of
ZmDof1 transcription factor in sorghum and wheat has activated the primary target, phospho
enolpyruvate carboxylase (PEPC), which leads to the down-regulation of genes involved in photosynthesis and the functional apparatus of chloroplasts, and negatively impacted photosynthesis, and biomass (
Peña et al., 2017). These results indicated that transcription factor strategies for crop improvement need to consider the downstream targets of the genetic elements to be introduced. Another category of regulatory genetic factors is stress signaling genes. Components of one signal transduction pathway may be shared by various stresses such as drought, salt and cold (Shinozaki and Yamaguchi-Shinozaki, 1999). Moreover, manipulation of signaling factors can control a broad range of downstream targets and result in superior tolerance to multiple stresses (Umezawa et al., 2006). Mitogen activated protein kinase (MAPK) cascade is an evolutionarily conserved signal transduction module involved in plant abiotic stress tolerance (Xiong and Zhu, 2001). The
NPK1 gene, a tobacco mitogen-activated protein kinase kinase kinase (MAPKKK), activated an oxidative signaling cascade and resulted in cold, heat, salinity and drought tolerant transgenic plants (Kovtun et al., 2000; Shou et al., 2004a).
Sorghum transformation and regeneration
To improve drought tolerance and validate the hypothesis that expression of NPK1 gene in sorghum would improve drought tolerance in sorghum, two genotypes: Dorado and SPGM94021 were transformed with the pSHX004 construct. To the best of our knowledge, this is the first report of transforming sorghum plants with the NPK1 gene. To select the transformed calli, the herbicide-resistant gene, bar (Thompson et al., 1987) was employed as a selectable marker. Selection of bialaphos-resistant callus is often conducted at 2 to 3 mg/L bialaphos (Casas et al., 1993; Frame et al., 2002; Grootboom et al., 2010). Bialaphos, as selection agent, has been found to hinder regeneration capacity of sorghum (Casas et al., 1993; Gao et al., 2005a; Lu et al., 2009; Grootboom et al., 2010). In the present investigation, bialaphos imposed a severe selection on sorghum callus. To sustain callus regenerability, bialaphos concentration was not increased beyond 1.5 mg/L. This is because non-transgenic plantlets may escape the bar selection system (Gao et al., 2005b).
Analysis of the transgenic plants
Further screening was conducted through PCR analysis, the transformation frequency was calculated and found to be within the cited range (Emani et al., 2002; Gao et al., 2005b; Zhao et al., 2000; Howe et al., 2006) from 0.2 to 4.5%, even though these reports were on highly regenerable and transformable genotypes, e.g. P898012 and TX430. The low transformation frequency may attribute to the detrimental effects of the NPK1 gene. In this respect, Kovtun et al. (1998) reported that overexpression of NPK1 gene causes detrimental effects on embryogenesis. Moreover, the constitutive expression of a calcium dependent protein kinases, OsCDPK-7, in sorghum induced apoptotic cell death in transgenic leaves (Mall et al., 2011). Further supporting this hypothesis is that the regeneration frequency for these genotypes is as high as 22 and 16% for Dorado and SPGM94021, respectively (Assem et al., 2014). Also, the Agrobacterium-mediated transformation frequency of Dorado with similar construct, pTF102, was as high as 3.6% (Zamzam, 2014). In this respect, slight reduction in the transformation frequency with the pSHX004 construct compared to pTF102 was also reported in maize (Shou et al., 2004b).
Taken together, the reduction in transformation frequency with pSHX004 construct in this study may be attributed to the death of the high NPK1 expressers during the different transformation stages. To further confirm the transgenic plants, Southern blotting was employed. Digesting the pSHX004 plasmid with the restriction enzyme EcoRI would liberate the NPK1 gene from the T-DNA giving one band at the size of 1.8 Kb. Different Southern hybridization patterns seen with EcoRI-digested DNA implied that the transgenic lines resulted from independent transformation events. The number of hybridization bands ranged from one to four. The presence of more than one band and of bands larger than 1.8 Kb may be attributed to the occurrence of rearrangement or to duplication or amplification of transgenes. Duplication or amplification of the transgene was reported by Spencer et al. (1992) in maize and Cannell et al. (1999) in wheat. The transgene copy number in the recipient genome is an important determinant of the transgene expression and field performance of the transgenic plants.
Agrobacterium is a potent transformation tool with the advantage of producing transgenic plants with low transgene copy number. In the present study, the low transgene copy number is consistent with other investigations using Agrobacterium on cereals (Hiei et al., 1994; Ishida et al., 1996; Cheng et al., 1997; Zhao et al., 2000; Gao et al., 2005a; Lu et al., 2009). The expression of the NPK1 transgene is the key of this investigation. Suppression and overexpression of NPK1 in tobacco have resulted in some detrimental effects on cell division, embryogenesis, and seed development (Kovtun et al., 1998). In this study, although no abnormal transgenic plants were observed, the transformation efficiency was rather low. It is likely that only transgenic sorghum events with relatively low expression of the transgene were generated in this study.
Although, quantitative real time PCR analysis revealed high expression of the NPK1 transgene in some T0 plants, that is, events No. 9, 12 and 14, low expression was found in other events (event No. 2 and 8). Transgenic event number 2 was almost silenced and it is likely that the T-DNA was inserted in a heterochromatin region of the chromosome and had low access to the RNA polymerase. In this respect, gene silencing has been previously reported in transgenic sorghum (Casas et al., 1993, 1997; Zhu et al., 1998; Able et al., 2001; Emani et al., 2002). Events 9 and 12 will be the main candidates for carrying out further investigation of the effects of overexpressing the NPK1 gene on field performance. On the other hand, examining the field performance of event 2 may shed the light on the effects of attenuating the expression of this gene.
Evaluation of T1 plants under drought stress conditions
The working hypothesis of this study was that expression of NPK1 gene in sorghum would improve drought stress tolerance. To investigate this hypothesis, transgenic plants were evaluated for growth and yield potential under drought stress. Mannitol has been used to evaluate plant tolerance to drought stress. Mannitol affects the availability of water to plants by increasing the osmotic pressure outside the cells and causes plant symptoms like that of water-deficit (Rumpho et al., 1983). In this investigation, the relative reduction in the growth parameters: fresh weight and shoot length of the non-transgenic seedlings, under mannitol stress, suggests that the presence of the NPK1 gene in transgenic sorghum plants aids in increasing the in vitro osmotic stress tolerance. The relative improvement in the growth of the transgenic plants, under mannitol stress, may be explained by the reduced sensitivity of transgenic cells to stress conditions. In this respect, Kovtun et al. (2000) reported that the ANP1, a class of MAPKKKs from Arabidopsis can be induced specifically by H2O2 and can activate a specific class of stress-induced MAPKs.
The activated MAPK cascade activates stress-response genes that protect plants from diverse environmental stresses. Moreover, Shou et al. (2004a) reported that the expression of NPK1 in transgenic maize enhanced drought tolerance, suggesting that NPK1 induced a mechanism that protected plants from dehydration damages. Similar results were, also, described, in Maize, by Muoma and Ombori (2014). Over-expression of NPK1 was found to impose detrimental effects on seed development in tobacco and the number of the defective seeds correlated with the level of the transgene expression, suggesting that seed phenotype was due to transgene expression and enhanced NPK1-dependent MAPK activity (Kovtun et al., 1998). In this study, we did not notice any reduction in seed germination or defect in seed development. Contrarily, under drought conditions, the transgenic plants had a higher means for grain weight despite the provision of equal amount of water. The relatively low expression of the NPK1 in this study may explain the normal development of seeds. The increased weight of transgenic kernels under drought condition in this study is consistent with the results of Shou et al. (2004a). Taken together, the results on the mannitol tolerance and the yield potential of transgenic sorghum plants expressing NPK1, demonstrate that NPK1 gene might play a role in the protection of plants under water deficiency- stress conditions. Therefore, the NPK1 gene can be successfully used for genetic improvement of Egyptian sorghum inbred lines for osmotic tolerance.
The authors have not declared any conflict interests.
This project was supported financially by Egypt’s Science and Technology Development Fund (STDF), Egypt. The binary vector pSHX004 has been kindly provided by The Plant Transformation Facility, Iowa State University, USA. Sorghum lines have been provided by Shandaweel Research Station, Sorghum Department, Field Crops Research Institute, ARC, Egypt.
REFERENCES
|
Able JA, Rathus C, Godwin ID (2001). The investigation of optimal bombardment parameters for transient and stable transgene expression in sorghum. In vitro Cell. Dev. Biol. Plant 37:341-348.
Crossref
|
|
|
|
Acquaah G (2007). Principles of Plant Genetics and Breeding. Wiley-Blackwell, Boston, Oxford, Chichester, Berlin, Singapore, Melbourne, Tokyo, Beijing.
|
|
|
|
|
Assefa Y, Staggenborg SA (2010). Grain sorghum yield with hybrid advancement and changes in agronomic practices from 1957 through 2008. Agron. J. 102:703-706.
Crossref
|
|
|
|
|
Assem SK, Zamzam MM, Hussein BA, Hussein EHA (2014). Evaluation of somatic embryogenesis and plant regeneration in tissue culture of ten sorghum (Sorghum bicolor L.) genotypes. Afr. J. Biotechnol. 13:3672-3681.
Crossref
|
|
|
|
|
Belton PS, Taylor JR (2004). Sorghum and millets: protein sources for Africa. Trends Food Sci. Technol. 15:94-98.
Crossref
|
|
|
|
|
Bhatnagar-Mathur P, Vadez V, Sharma KK (2008). Transgenic approaches for abiotic stress tolerance in plants: retrospect and prospects. Plant Cell Rep. 27:411-424.
Crossref
|
|
|
|
|
Cannell ME, Doherty A, Lazzeri PA, Barcelo P (1999). A population of wheat and tritordeum transformants showing a high degree of marker gene stability and heritability. Theor. Appl. Genet. 99:772-784.
Crossref
|
|
|
|
|
Casas AM, Kononowicz AK, Haan TG, Zhang L, Tomes DT, Bressan RA, Hasegawa PM (1997). Transgenic sorghum plants obtained after microprojectile bombardment of immature inflorescences. In Vitro Cell. Dev. Biol. Plant 33:92-100.
Crossref
|
|
|
|
|
Casas A.M., Kononowicz A.K., Zehr UB, Tomes DT, Axtell JD, Butler LG, Bressan RA, Hasegawa P (1993). Transgenic sorghum plants via microprojectile bombardment. Proc. Natl. Acad. Sci. USA. 90:11212-11216.
Crossref
|
|
|
|
|
Cheng M, Fry JE, Pang S, Zhou H, Hironaka CM, Duncan DR, Conner TW, Wan Y (1997). Genetic transformation of wheat mediated by Agrobacterium tumefaciens. Plant Physiol. 115:971-980.
Crossref
|
|
|
|
|
Das K. and Roychoudhury A. (2014). Reactive oxygen species (ROS) and response of antioxidants as ROS-scavengers during environmental stress in plants. Frontiers Environ. Sci. 2(53):1-13.
Crossref
|
|
|
|
|
Devi P, Zhong H, Sticklen M (2004). Production of transgenic sorghum plants with related HVA1 gene. In: Seetharama N, Godwin ID. (eds), Sorghum Tissue Culture and Transformation. Oxford & IBH Publishing Co. Pvt. Ltd, New Delhi.
|
|
|
|
|
Emani C, Sunilkumar G, Rathore KS (2002). Transgene silencing and reactivation in sorghum. Plant Sci. 2:181-192.
Crossref
|
|
|
|
|
Frame BR, Shou H, Chikwamba RK, Zhang Z, Xiang C, Fonger TM, Pegg SE, Li B, Nettleton DS, Pei D, Wang K (2002). Agrobacterium tumefaciens-mediated transformation of maize embryos using a standard binary vector system. Plant Physiol. 129:13-22.
Crossref
|
|
|
|
|
Gao Z, Jayaraj J, Muthukrishnan S, Claflin L, Liang GH (2005a). Efficient genetic transformation of sorghum using a visual screening marker. Genome 48:321-333.
Crossref
|
|
|
|
|
Gao Z, Xie X, Ling Y, Muthukrishnan S, Liang GH (2005b). Agrobacterium tumefaciens-mediated sorghum transformation using a mannose selection system. Plant Biotechnol. J. 3: 591-599.
Crossref
|
|
|
|
|
Grootboom AW, Mkhonza NL, O`Kennedy MM, Chakauya E., Kunert K, Chikwamba R K (2010). Biolistic Mediated Sorghum (Sorghum bicolor L. Moench) Transformation via Mannose and Bialaphos Based Selection Systems. Int. J. Bot. 6: 89-94.
Crossref
|
|
|
|
|
Hiei Y, Ohta S, Komari T, Kumashiro T (1994). Efficient transformation of rice (Oryza sativa L.) mediated by Agrobacterium and sequence analysis of the boundaries of the T-DNA. Plant J. 6:271-282.
Crossref
|
|
|
|
|
Horwich L. (2014). Molecular Chaperones in Cellular Protein Folding: The Birth of a Field. Cell 157:285-288.
Crossref
|
|
|
|
|
Howe A, Sato S, Dweikat I, Fromm M, Clemente T (2006). Rapid and reproducible Agrobacterium-mediated transformation of sorghum. Plant Cell Rep. 25:784-791.
Crossref
|
|
|
|
|
Ishida Y, Saito H, Ohta S, Hiei Y, Komari T, Kumashiro T (1996). High efficiency transformation of maize (Zea mays L.) mediated by Agrobacterium tumefaciens. Nat. Biotechnol. 14:745-750.
Crossref
|
|
|
|
|
Kasuga M, Liu Q, Miura S, Yamaguchi-Shinozaki K, Shinozaki K (1999). Improving plant drought, salt, and freezing tolerance by gene transfer of a single stress inducible transcription factor. Nat. Biotechnol. 17:287-291.
Crossref
|
|
|
|
|
Kim H, Lee K, Hwang H, Bhatnagar N, Kim DY, Yoon IS, Byun MO, Kim ST, Jung KH, Kim BG (2014). Overexpression of PYL5 in rice enhances drought tolerance, inhibits growth, and modulates gene expression. J. Exp. Bot. 65:453-464.
Crossref
|
|
|
|
|
Kovtun Y, Chiu WL, Tena G, Sheen J (2000). Functional analysis of oxidative stress-activated mitogen- activated protein kinase cascade in plants. Proc. Natl. Acad. Sci. USA 97:2940-2945.
Crossref
|
|
|
|
|
Kovtun Y, Chiu WL, Zeng W, & Sheen J. (1998). Suppression of auxin signal transduction by a MAPK cascade in higher plants. Nature 395:716-720.
Crossref
|
|
|
|
|
Li HS, Chang CS, Lu LS, Liu CA, Chan MT, Charng YY (2003). Over-expression of Arabidopsis thaliana heat shock factor gene (AtHsfA1b) enhances chilling tolerance in transgenic tomato. Bot. Bull. Acad. Sin. 44:129-140.
|
|
|
|
|
Lu L, Wu X, Yin X, Morrand J, Chen X, Folk WR, Zhang ZJ (2009). Development of marker-free transgenic sorghum [Sorghum bicolor (L.) Moench] using standard binary vectors with bar as a selectable marker. Plant Cell Tiss. Organ Cult. 99: 97-108.
Crossref
|
|
|
|
|
Maheswari M, Varalaxmi Y, Vijayalakshmi A, Yadav SK, Sharmila P, Venkateswarlu B, Vanaja M, Saradhi PP (2010). Metabolic engineering using mtlD gene enhances tolerance to water deficit and salinity in sorghum. Biol. Plantarum 54: 647-652.
Crossref
|
|
|
|
|
Mall TK, Dweikat I, Sato SJ, Neresian N, Xu K, Ge Z, Wang D, Elthon T, Clemente T (2011). Expression of the rice CDPK-7 in sorghum: molecular and phenotypic analyses. Plant Mol. Biol. 75:467-479.
Crossref
|
|
|
|
|
Milesi C, Samanta A, Hashimoto H, Kumar KK, Ganguly S, Thenkabail P.S., Srivastava AN, Nemani RR, Myneni RB (2010). Decadal Variations in NDVI and Food Production in India. Remote Sens. 2:758-776.
Crossref
|
|
|
|
|
Mortimore M (2009). Dryland Opportunities: A New Paradigm for People, Ecosystems and Development. IUCN, Gland, Switzerland, IIED, London, UK, UNDP, New York, USA.
|
|
|
|
|
Muoma JVO, Ombori O (2014). Agrobacterium-Mediated Transformation of Selected Kenyan Maize (Zea mays L.) Genotypes by Introgression of Nicotiana Protein Kinase (npk1) to Enhance Drought Tolerance. Am. J. Plant Sci. 5:863-883.
Crossref
|
|
|
|
|
Murray MG, Thompson WF (1980). Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res. 8:4321-4325.
Crossref
|
|
|
|
|
Pe-a PA, Quach T, Sato S, Ge Z, Nersesian N, Changa T, Dweikat I, Soundararajan M, Clemente TE (2017). Expression of the Maize Dof1 Transcription Factor in Wheat and Sorghum. Front. Plant Sci. 8:434-447.
|
|
|
|
|
Rumpho ME, Edwards GE, Loescher WH (1983). A pathway for photosynthetic carbon flow to mannitol in celery leaves: activity and localization of key enzymes. Plant Physiol. 73:869-873.
Crossref
|
|
|
|
|
Shinozaki K, Yamaguchi-Shinozaki K (1999). Molecular responses to drought stress. In: Shinozaki K. & Yamaguchi-Shinozaki K. (eds), Molecular Responses to Cold, Drought, Heat and Salt Stress in Higher Plants. RG Landes Co, Austin.
|
|
|
|
|
Shou H, Bordallo P, Wang K (2004a). Expression of the Nicotiana protein kinase (NPK1) enhanced drought tolerance in transgenic maize. J. Exp. Bot. 55:1013-1019.
Crossref
|
|
|
|
|
Shou H, Bordallo P, Fan JB, Yeakley JM, Bibikova M, Sheen J, Wang K (2004b). Expression of an active tobacco mitogen-activated protein kinase kinase kinase enhances freezing tolerance in transgenic maize. Proc. Natl. Acad. Sci. USA 101:3298-3303.
Crossref
|
|
|
|
|
Spencer TM, O'Brien JV, Start WG, Adams TR, Gordon-Kamm WJ & Lemaux, PG (1992). Segregation of transgenes in maize. Plant Mol. Biol. 18:201-210.
Crossref
|
|
|
|
|
Thompson CJ, Movva NR, Tizard R, Crameri R, Davies JE, Lauwereys M, Botterman J (1987). Characterization of the herbicide-resistance gene bar from Streptomyces hygroscopicus. EMBO J. 6:2519-2523.
|
|
|
|
|
Tuberosa R, Salvi S (2006). Genomics-based approaches to improve drought tolerance of crops. Trends Plant Sci. 11:405-412.
Crossref
|
|
|
|
|
Umezawa T, Fujita M, Fujita Y, Yamaguchi-Shinozaki K, Shinozaki K. (2006). Engineering drought tolerance in plants: discovering and tailoring genes to unlock the future. Curr. Opin. Biotechnol. 17:113-122.
Crossref
|
|
|
|
|
Wenzel WG (1999). Effect of moisture stress on sorghum yield and its components. South Afr. J. Plant Soil 16:153-157.
Crossref
|
|
|
|
|
Wilhite DA (2000). Drought as a natural hazard in concepts and definitions. In: Wilhite D.A. (ed), Drought: A Global Assessment. Routledge, Oxfordshire, New York City, Melbourne, New Delhi, Singapore, Beijing.
|
|
|
|
|
Xiong L, Zhu JK (2001). Abiotic stress signal transduction: molecular and genetic perspectives. Physiol. Plantarum 112:152-166.
Crossref
|
|
|
|
|
Yellisetty V, Reddy LA, Mandapaka M (2015). In planta transformation of sorghum (Sorghum bicolor (L.) Moench) using TPS1 gene for enhancing tolerance to abiotic stresses. J. Genet. 94:425-434.
Crossref
|
|
|
|
|
Zamzam MM (2014). Genetic Improvement of Some Sorghum Lines via Genetic Engineering Approaches. M.Sc. Thesis, Faculty of Agriculture, Cairo University, Egypt. 170p.
|
|
|
|
|
Zhao Z, Cai T, Tagliani L, Miller M, Wang N, Pang H, Rudert M, Schroeder S, Hondred D, Seltzer J, Pierce D (2000). Agrobacterium-mediated sorghum transformation. Plant Mol. Biol. 44:789-798.
Crossref
|
|
|
|
|
Zhu H, Muthukrishnan S, Krishnaveni S., Jeoung JM, Liang, GH (1998). Biolistic transformation of sorghum using a rice chitinase gene. J. Genet. Breed. 52:243-252.
|
|