Exploration of efficacious plant extracts that can reduce or inhibit Pseudomonas aeruginosa biofilm formation is necessary. Allium sativum is a suitable candidate because of its relative abundance. This study was carried out to determine the effect of ethanolic A. sativum extract on the expression of the P. aeruginosa biofilm gene, pelF. The presence of the pelF gene in the isolates used for this study was confirmed via polymerase chain reaction (PCR) and agarose gel electrophoresis. P. aeruginosa cultures treated with 1 g/ml of the A. sativum extract had the least turbidity (6.7 absorbance value at A600). The expression profile of the pelF gene in the treated cultures was determined via PCR using cDNA synthesized from RNA isolated from the treated P. aeruginosa cultures. The amplicons from the PCR were analyzed via agarose gel electrophoresis and a concentration dependent down-regulation of the pelF gene was observed. Further quantification of the pelF gene’s expression was performed via real-time PCR using rpoB as the reference gene. A 4-fold down-regulation of the gene was observed at 0.5 and 1 g/ml concentrations, respectively. This study suggests that the suppression of the pelF gene of P. aeruginosa by A. sativum extracts plays a role in the inhibition of P. aeruginosa biofilm formation. This is the first study to elucidate the effect of A. sativum on the expression of any of the pel genes.
Several cases of Pseudomonas aeruginosa infections have been reported which implicate the organism as a leading cause of nosocomial infections. An estimated 10% of all nosocomial infections are caused by P.
aeruginosa, especially in immuno-compromised individuals (Aloush et al., 2006; Lund-Palau et al., 2016). Keratitis, pneumonia, scleritis, cystic fibrosis, urinary tract infections, wound infections resulting from invasive and surgical procedures, open and burn wounds are usually caused by P. aeruginosa (although, often in association with other bacteria) (Okamoto et al., 2001; Fernandez-Barat et al., 2017; Murugan et al., 2017).
The virulence of P. aeruginosa has been shown to depend on the expression of genes that encode its virulence factors. These virulence factors are classified based on the type of infections caused (chronic or acute infections). Virulence factors expressed during chronic infections include biofilms, alginate, elastase secretion, acyltransferase, pyoverdin and pyochelin (Lang et al., 2016). Virulence factors associated with acute P. aeruginosa infections include exotoxin A, pili, phospholipase C and exoenzyme S (Faraji et al., 2016; Valadbeigi et al., 2017).
P. aeruginosa infections are difficult to treat due to various mechanisms that confer on it intrinsic resistance to common antibiotics (Tanya and Daniel, 2009; Khalaji et al., 2013). The inefficiency of common antimicrobials to treat infections makes it necessary for plant candidates to be exploited in achieving prophylaxis. Hyper-virulent strains which demonstrate susceptibility to antibiotics still cause difficult-to-treat infections in patients. Murugan et al. (2017) studied patients with scleritis caused by P. aeruginosa VRFPA10. Their analyses of the P. aeruginosa VRFPA10 genome showed genomic islands which they attributed the hyper-virulence to.
Biofilm formation in P. aeruginosa, a quorum-sensing controlled mechanism, has been reported to increase its resistance to antibiotics 1000-fold. The las quorum sensing system of P. aeruginosa controls the transcription of the pel gene operon which consists of the following genes: pelA, pelB, pelC, pelD, pelE, pelF and pelG (Bacalso et al., 2011; Xu et al., 2013). The pel genes (pellicle biosynthetic genes) have been shown to code for proteins involved in the synthesis of the exopolysaccharide moieties during P. aeruginosa biofilm formation (Vasseur et al., 2005; Sakuragi and Kolter, 2007). The roles of the individual pel genes in the Pel biosynthetic pathway have not been fully established. The pelF gene however, has been one of the most extensively studied and it encodes a glycosyltransferase. It is the only gene in the operon that produces a protein that localizes in the cytoplasm (Vasseur et al., 2005; Franklin et al., 2011).
Several plant extracts have been studied for their quorum sensing (QS) inhibitory properties (Singh et al., 2012). In this study,
Allium sativum (garlic) extract was used to treat
P. aeruginosa isolated from wounds. Garlic has been reported to inhibit quorum sensing in
P. aeruginosa, thereby, allowing better penetration of antimicrobials into the biofilm matrix (Cavallito and Bailey, 1944; Hurley et al., 2012; Harjai et al., 2009). Studies have also demonstrated the ability of garlic extracts to inhibit biofilm formation in planktonic bacteria (Arzanlou et al. 2007;
Mohsenipour and
Hassanshahian, 2015). Being a plant that has been widely used for its medicinal properties, bacteria are unlikely to develop resistance to garlic extracts. This study seeks to determine the effects of
A. sativum on the expression of the
P. aeruginosa biofilm gene,
pelF. Also, this study seeks to establish the prospect for use of garlic extracts either in individual or combined treatment of wound infections by
P. aeruginosa.
Bacterial isolation
P. aeruginosa were isolated from 10 wound swabs collected from patients in the medical ward of Barau Dikko Teaching Hospital Kaduna (this was done with the approval of the hospital’s management). Fresh garlic bulbs were purchased from the Central Market, Kaduna, Nigeria. This study was carried out in the Microbiology Laboratory of Kaduna State University, Nigeria.
Cetrimide agar (Merck, Germany) was prepared according to the manufacturer’s instructions. Briefly, 45 g of the agar powder was weighed and dissolved 1 L of distilled water by heating at 50°C in a water bath. Then, 3 ml of glycerol (Merck, Germany) was added and the mixture was autoclaved at 121°C for 15 min. Sterilized media was aseptically poured into sterile petri dishes, covered and allowed to solidify. The wound swabs were aseptically streaked on the solidified media and incubated for 24 h at 37°C.
Exactly two drops of the oxidase reagent (Dalynn, Canada) were used to moisten a piece of filter paper. Greenish colonies from the 24 h culture were smeared on the moistened filter paper using an applicator stick. Colour change was observed within 30 s (
Cheesbrough, 2000).
Preparation of the ethanolic extract of A. sativum
Fresh whole bulbs of A. sativum (garlic) were washed with distilled water and the outer cover was removed using a sharp knife disinfected with 75% ethanol. Exactly, 500 g of the washed garlic was cut into pieces and was ground using a disinfected blender; it was then transferred to a sterile beaker containing 100 ml of 70% ethanol. The beaker was covered and left to stand for 24 h at room temperature. The mixture was then filtered using Whatman No 1 filter paper. The filtrate was poured into a crucible and placed in a water bath to concentrate at 50°C. The filtrate was exposed to the air to allow the ethanol to evaporate leaving only the pelleted A. sativum (A. sativum) extract.
To prepare various concentrations of the A. sativum extracts, the pellets were crushed into fine powder using a mortar and pestle. Then, 10, 5, 3.3, 2.5 and 2 g of the powder were each dissolved in 10 mL of distilled water to obtain the following concentrations of the extract (g/mL): 10, 5, 3.3, 2.5 and 2.
Preparation of broth cultures of P. aeruginosa
Nutrient broth (Merck, Malaysia) was prepared according to the manufacturer’s instructions. Briefly, 1.2 g of the powder was dissolved in 150 ml of distilled water by swirling. The dissolved broth was autoclaved at 121°C for 15 min.
To prepare the broth culture of the test organism, 10 ml of the nutrient brought was poured into a test tube. A single colony of the P. aeruginosa isolates was transferred to the nutrient broth using a sterile wire loop and mixed. It was incubated at 37°C with shaking at 200 rpm until the culture turbidity matches 0.5 McFarland standard (corresponding to approximately 1.5 X 108 CFU/ml) (Ndip et al., 2005).
Detection of the pelF and rpoB genes
To proceed with this study, it was important to detect the presence of the pelF gene in the representative P. aeruginosa isolate so as to justify the proceeding gene expression study. The presence of the rpoB gene, which was used as the reference gene (housekeeping gene) in quantifying the fold change in gene expression was also detected.
Specific pair of primers for the P. aeruginosa pelF and rpoB genes were designed using the online primer design tool, NCBI primer BLAST. The specificity of the designed primers to the pelFand rpoB gene sequences was confirmed using the online tool IDT OligoAnalyzer 3.1.
Probing for, and the amplification of the pelF and rpoB genes were carried out in the P. aeruginosa isolate employed in this study using the polymerase chain reaction technique (PCR) as described by Gemiarto et al. (2015). This was performed to ascertain the presence of the genes in the test organism. Genomic DNA was isolated from the P. aeruginosa culture using phenol-chloroform extraction method. Purity of the extracted DNA was ascertained spectrophotometrically using the A260/A280 ratio.
Exactly, 2 µl of the extracted P. aeruginosa genomic DNA was added to 12.5 µl of 2X GoTaq Green PCR Master Mix (Promega, USA), 0.4 µl each of the appropriate forward and reverse primers designed using the NCBI primer BLAST (Basic Local Alignment Search Tool) and nuclease-free water to a final volume of 25 µl and mixed gently.
The PCR was carried out in a thermocycler (Applied Biosystems, USA). An initial denaturation at 95°C for 10 min was carried out followed by 30 cycles of denaturation at 94°C for 30 s, annealing temperature of 55°C for 45 s and elongation at 72°C for 45 s; then a final elongation at 72°C for 5 min was done. Analysis of the PCR products was carried out via agarose gel electrophoresis (using 1% agarose in TAE buffer and ethidium bromide dye).
After the PCR amplification, the suspected DNA amplicons (pelF and rpoB) were separated using agarose gel electrophoresis. This was followed by the gel extraction of the obtained DNA bands suspected to be those of pelF and rpoB gene from the agarose gel and subjecting same to sequencing. The degree of alignment of the sequenced DNA to those of the pelF and rpoB genes determined in the NCBI gene bank was obtained using the NCBI BLAST.
Treatment with various concentrations of A. sativum extract
For each concentration of the extract, 8 ml of freshly prepared nutrient broth was poured into a test tube and 1 ml of the P. aeruginosa broth cultured was added to it. Then, 1 ml each of the various concentrations (10, 5, 3.3, 2.5 and 2 g/ml) of the A. sativum extract was added to the respective test tubes. For each concentration, the treatment was carried out in triplicates. The treated broth cultures including an untreated control, were incubated in 100 ml conical flasks (to provide a greater surface area for attachment and biofilm formation), at 37°C for 24 h.
The overnight cultures of the treated cells and the untreated control were sub-cultured on cetrimide agar to compare the cell densities. This is to check for viability of the cells after the treatment.
Turbidity measurement of treated cultures
The turbidity of the treated P. aeruginosa cultures was determined using a spectrometer (Labomed, USA). Briefly, sterile swab sticks were used to gently dislodge cells that were attached to the bottom of the conical flask. The culture was mixed by swirling for 1 min. Then, about 2 ml of each of the treated cultures (and the untreated control) was transferred into a quartz cuvette. The measurements were taken at A600.
Analysis of pelF gene expression following treatment with A. sativum extract
Total RNA from the P. aeruginosa cultures treated with the A. sativum extract was isolated according to the manufacturer’s instructions, using the SV Total RNA Isolation System (Promega, USA). Purity of the extracted RNA was ascertained spectrophotometrically using the A260/A280 ratio. The isolated RNA was reverse-transcribed to synthesize cDNA, according to the manufacturer’s instructions, using the AccuPower RT-PCR premix (Bioneer, USA).
The synthesized cDNA were analyzed on 1% agarose gel to determine if the pelF gene was expressed. This step is necessary before carrying out qPCR to quantify the gene expression.
Real-time quantitative PCR for differential pelF gene expression
Quantitative PCR (qPCR) was carried out to ascertain the effect of the ethanolic A. sativum extract on the pelF gene. The rpoB gene was used as the housekeeping gene and the cDNA synthesized in the section above was used as the template. The qPCR was performed using the 2X GoTaq qPCR mastermix (Promega, USA) and the reaction was set up according to the manufacturer’s protocol. The ABI StepOne (Applied Biosystems, USA) real-time thermal cycler was used and the cycling conditions used are as follows: heat activation at 95°C for 2 min then 40 cycles of denaturation (95°C for 15 s), annealing (57°C for 30 s) and extension (60°C for 60 s). A final denaturation from 60 to 95°C for 30 s was carried out. Quantification of the fold change in gene expression was calculated using the formula employed in the Pfaffl (2001) method.
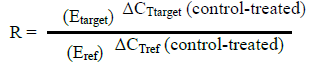
Where, R = ratio; E = efficiency of the primers; ΔCT = difference between CT (cycle threshold) of control and treated samples; Ref = reference gene (rpoB); target = the amplified gene (pelF gene).
The cDNA that was reverse-transcribed from the untreated P. aeruginosa was used to generate five-fold serial dilutions for the derivation of a standard curve. Differential expression of the pelF gene was derived by comparing CT (cycle threshold) values obtained from it with those of the housekeeping gene used (rpoB).
Changes in gene expression are only considered to be significant when the fold change is ≥2 (where the gene is said to be up -regulated) or ≤-2 (where the gene is said to be down-regulated).
Isolation and identification of P. aeruginosa
Of the 10 wound swab samples collected, 5 produced green colonies when cultured on cetrimide agar (a selective medium for P.aeruginosa), and they were confirmed to be P. aeruginosa by the production of an intense purple color during the oxidase test.
Detection of the pelF and rpoB genes in the isolates
Following the isolation of DNA from the isolate, pairs of specific primers (Table 1) were used to amplify the pelF and the rpoB genes and the amplicons were analyzed via agarose gel electrophoresis using 1% agarose in TAE buffer and ethidium bromide dye. Amplicon sizes corresponding to the expected product size of the primers used (452 and 1382 bp respectively) were obtained. The results obtained are as shown in Figure 1a and b.
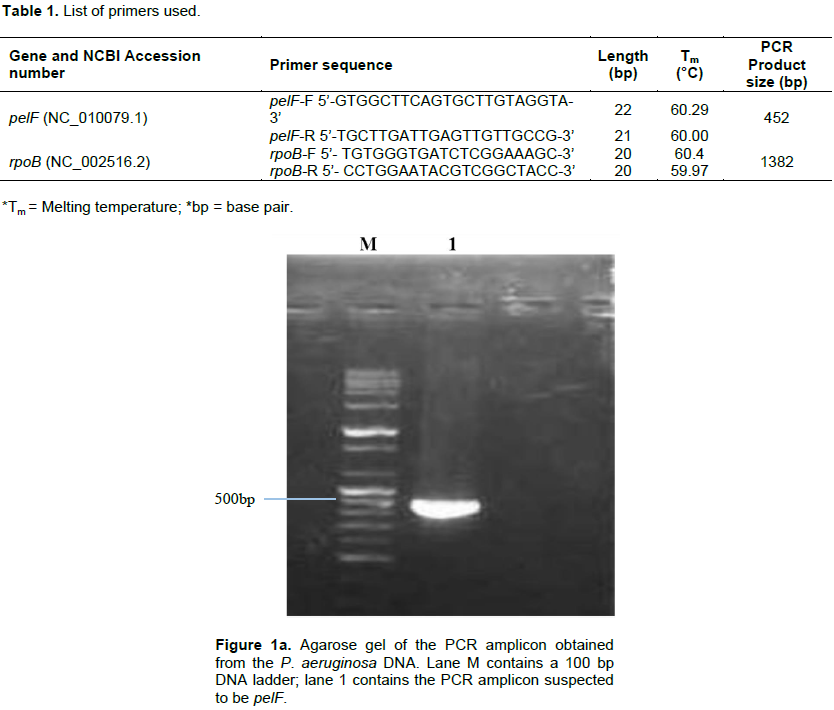
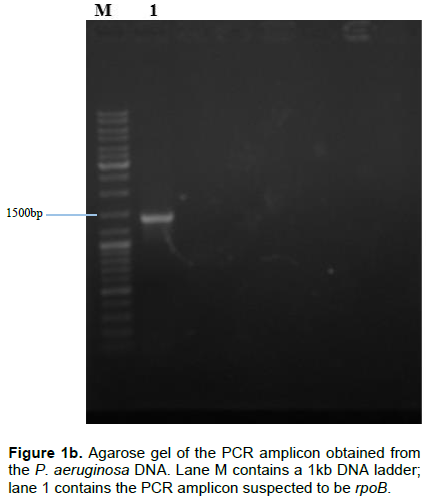
Pel, produced by P. aeruginosa has been reported to also serve the function of preserving the capacity of the organism to continue to produce biofilms even when other exopolysaccharide genes have undergone mutation (Colvin et al., 2012). This validates the choice of the pel genes as a suitable candidate for down-regulating biofilm production in P. aeruginosa. In all the P. aeruginosa isolated from the wound swabs collected for this study, pelF was detected. This suggests that this QS-regulated gene is ubiquitous and highly conserved (Bacalso et al., 2011). The rpoB, which is responsible for the synthesis of the beta subunit of the RNA polymerase is an important reference gene for qPCR because it is expressed at a consistent level as long as the cell is alive.
Treatment of P. aeruginosa broth cultures with various concentrations of A. sativum extract
The various broth cultures of the test organism for this study were treated with 1, 0.5, 0.33, 0.25, 0.20 and 0 g/ml concentrations of the ethanolic A. sativum extract.Absorbance readings of the treated cultures at A600, following overnight incubation showed that: with increasing concentration of the extract, a decrease in the turbidity of the treated cultures was observed. This was recorded as an increase in the amount of light that passes through the culture; the less the turbidity, the more light passes through it. The culture treated with 1 g/ml of the extract had the least turbidity (6.7 absorbance value) and that treated with 0.20 g/ml of the extract had the highest turbidity (2.0 absorbance value) (as shown in Figure 2).
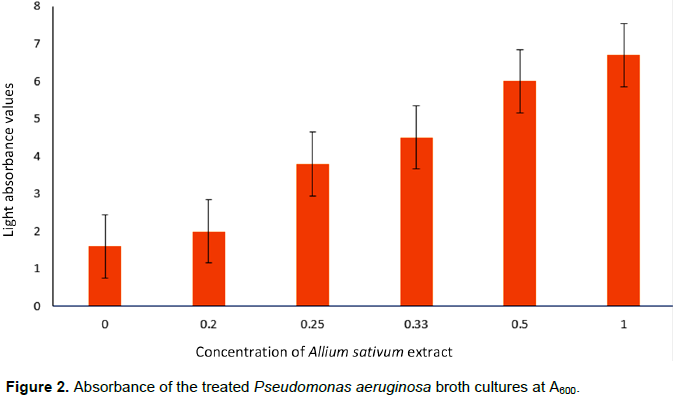
Figure 2 shows that the concentration of the A. sativum applied is inversely proportional to the intensity of the turbidity of the culture and the highest turbidity (least light absorption) was observed in the untreated control (absorbance value: 1.6) and highest when treated with the 1 g/ml extract concentration (6.7 absorbance value). The intensity of the culture turbidity observed is dependent on amount of cells that form the biofilm at the base of the culture flask used. As the concentration of the extract was increased, the amount of cells layered at the base of the flask decreased. This clearly shows that the decreasing turbidity was concentration dependent. This also does not agree with earlier findings which reported that garlic extract increases biofilm formation by S. mutans (Lee et al., 2011).
In this study, the broth treated with the highest concentration (1 g/ml) of A. sativum extract has the highest absorbance value (6.7) and the broth treated with the lowest concentration had the lowest absorbance value (2.0). The rapid drop in the turbidity observed between concentrations 0.5 and 0.33 g/ml may imply that the minimum effective dose of the active component of the extract lies within that concentration range.
Determination of the expression profile of the pelF gene in the treated cultures
RNA (which is the first product of gene expression before its translation into proteins) was isolated from the P. aeruginosa cultures that were treated with the various concentrations of the ethanolic A. sativum extract. The isolated RNA, after being reverse transcribed into cDNA and amplified using a conventional PCR thermocycler; was analyzed via agarose gel electrophoresis using 1% agarose in TAE buffer and ethidium bromide dye. A down-regulation of the pelF gene was observed and estimated as the increasing faintness of the amplicon bands obtained on the agarose gel with increasing concentration of the extract used in the treatment of the broth cultures (as shown in Figure 3).
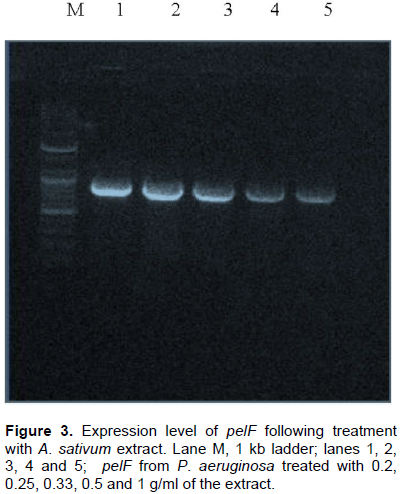
Figure 3 clearly shows a consistent decrease in the expression of the quorum sensing controlled gene, pelF, with increasing concentration of the extract. A study suggested that the thiosulfinates contained in garlic (A. sativum) is responsible for quorum sensing inhibition in P. aeruginosa (Vadekeetil et al., 2014). There are various
processes through which quorum sensing is inhibited (Gemiarto et al., 2015) and A. sativum-induced QS inhibition may be associated with the degradation of the signalling molecule (Vadekeetil et al., 2014).
To confirm that this increasing faintness in band quality is as a result of the down-regulation of the pelF gene and not due to cell death, the treated cultures were cultured on cetrimide agar plate and the growth obtained was compared against an untreated control. The growth obtained suggested that the test organism was still viable after treatment with the aforementioned concentrations of the A. sativum extract (data, not published).
Real-time quantitative PCR for differential pelF gene expression
Figure 4 shows that at extract concentrations of 0.2-0.33 g/ml, there was no significant up-regulation or down-regulation of the pelF gene. This could be due to the fact that these concentrations were not high enough to cause any fold-changes in the gene expression. At higher concentrations (0.5 and 1 g/ml), the A. sativum extract resulted in over 4-fold down-regulation of the pelF gene. Since the exact mechanism through which the expression of the pel genes are regulated is yet unknown (Sakuragi and Kolter, 2007), the role of the A. sativum extract in the down-regulation of the pelF gene cannot be inferred with precision.
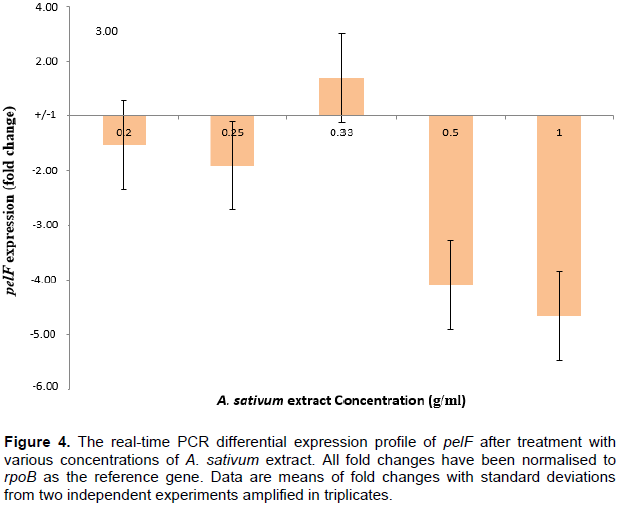
In an inducible gene operon, the expression of genes located downstream in the operon is usually regulated by the products synthesized by genes located closer to the promoter. Hence, the garlic extracts used in this study are very likely to have interacted with products synthesized by genes located from pelA to pelE, thereby lowering the induction of pelf, the result of which may be observed as the decreased transcription of the gene and the observed down-regulation. One clear fact is that this down-regulation of the pelF gene must have occurred before transcription of the gene; this conclusion is made because qPCR cannot measure post-transcriptional control of gene expression.
On a general note, the quorum sensing inhibitory properties of
A. sativum have been extensively elucidated. The organosulphur compound, allicin, found in garlic has been shown to act as a QS inhibitor (Bodini et al., 2009; Smyth et al., 2010). Hence, to confirm that the decrease in culture turbidity shown in Figure 2 and the expression of
pelF shown in Figure 3 was not as a result of cell death but as a decline in biofilm formation, the treated cultures were sub-cultured and colonies were observed. The colony count observed confirms that the
P. aeruginosa were not killed by the concentrations of the extract applied. This is in agreement with results from studies in which
P. aeruginosa was treated with natural organo-sulfur compounds as found in garlic (Arzanlou et al., 2007; Cady et al., 2012; Ratthawongjirakul and Thongkerd, 2016). They also observed no reduction in planktonic growth after treatment but they observed a decline in biofilm formation. A reduction in biofilm formation makes
P. aeruginosa more susceptible to antibiotics (Bjarnsholt et al., 2005; Indu et al., 2006). Hence, this finding could be exploited in using
A. sativum extracts in combination with antibiotics or other plant extracts for more effective antibiotic therapy. The prospects for achieving this is clearly reflected in the inhibition of
P. aeruginosa biofilm formation when a blend of essential oils from
Cinnamomum cassia,
A. sativum and
Mentha piperita were used to treat planktonic
P. aeruginosa PA01 strains (Lang et al., 2016)
.
This study could be extended to explore the gene expression of other pel genes in response to treatment with A. sativum extracts. Components of the extract employed here could be used individually to assess their effect on the expression of the P. aeruginosa pel genes. This study shows that increasing concentrations of A. sativum resulted in an increasing down-regulation of an important P. aeruginosa biofilm gene, pelF. This down-regulation was not due to extract-induced death of planktonic cells. This study supports the prospects for the use of A. sativum extracts as an antipathogenic remedy in combined therapy with antibiotics against resistant bacteria.