Full Length Research Paper
ABSTRACT
Escherichia coli multi-resistance to a variety of antimicrobials is a result of gene mutation on plasmids, integrons and transposons. The aims of this work were to: (1) detect genotype and phenotype antibiotic resistance genes in E. coli, and (2) determine whole-genome sequencing to discover E. coli gene multi-drug resistance in chicken meat. Samples were gathered, processed, and analysed bacteriologically; thereafter an antimicrobial sensitivity test was performed and E. coli isolates were identified serologically. Results of E. coli were 40% from 100 chicken samples. The most potent antibiotics against E. coli were Cephalosporins, Quinolones and Oxytetracycline. The serological investigation was as follows: 30% (O157:H7) of STEC, 30% (O142) of ETEC, 10% (O26:H11) of EHEC and 10% EPEC. Subunit B of Shiga-like toxin (SLT) gene showed a symmetrical band, while, Heat-labile toxin (LT) gene was estimated in both plasmid preps in addition to DNA genomic strains. STEC is hazardous to the chicken meat consumers. The study recommended necessary improvement in the hygienic procedures during all processing steps, and minimized the non-important usage of antibiotics to prevent antibiotics resistant.
Key words: Integrons, Shiga toxin-producing Escherichia coli, Gentamicin, heat-labile toxin, plasmids.
INTRODUCTION
Even though Escherichia coli is a nonpathogenic gram-negative intestinal microorganism, it is considered a commensal for both humans and animals. Enteropathogenic, Enteroinvasive, Enterotoxigenic and Enterohaemorrhagic are the four classifications of E. coli. However, infectious Shiga toxin-producing E. coli (STEC) can cause gastrointestinal disorders such as hemorrhagic colitis (HC), diarrhea and (HUS) haemolytic-uraemic syndrome by drinking contaminated water or eating infected food (Adeyanju and Ishola, 2014).
STEC is the more commonly investigated and serious pathogenic microorganisms that infect humans through poultry meat and are found after drinking contaminated water, dirty food, etc. Whereas beef, chicken and mutton are commonly contaminated by the fecal gut contents of the chicken itself during processing such as slaughter, or by consumers’ handlers during processing and storage (EL-Kholy et al., 2020).
E. coli infections are commonly treated by antibiotics. This results in presence of antibiotic resistant E. coli strain, making the microbe multidrug resistant to many antibacterial agents. Microorganisms have antibacterial resistance mainly due to gene mutation, and resistance genes exist in transposons, plasmids and integrons.
An integron is a two component gene capture and dissemination system, initially discovered in relation to antibiotic resistance, and which is found in plasmids, chromosomes and transposons. Integron Class I consists of: Two conversational segments (CS) (CDC, 2019) that are adjacent to the variable region (VR) and are common to the new generation of Enterobacteriaceae. Anti-bacterial resistance is due to frequent use of antibiotics resulting in a mutated generation of microbial strains, which increases resistant against various antibiotics. It is considered one of the most important public health crises in the world. Scientists are searching for a novel generation of drugs, which are more effective than the currently used resistant-type antibiotics that are used to treat diseased human, poultry and animals (Cunrath et al., 2019).
The purpose of the research is to 1) estimate genotype and phenotype of resistance antibiotic in E. coli and 2) identify the microbial resistance gene structure in whole-genome sequencing against poly resistant E. coli in commercially available poultry meat.a
MATERIALS AND METHODS
Sampling, preparation, bacterial testing
Approximately 100 chicken samples tested were randomly picked from various markets, stored in polyethylene bags, and then immediately transferred to a refrigerator bacteriological laboratory for analysis. Two grams of homogenized chicken meat sample was cultured in MacConkey broth and then incubated for 18 h at 37°C. Next, it was streaked into a MacConkey agar medium (Oxoid) plate at approximately 24 h/37°C. The pink colonies were drawn on eosin methylene blue (EMB) (Oxoid) media for approximately 24 h/37°C. The morphology of E. coli appeared as large colonies with a blue-black-green metallic luster. E. coli colonies were identified by morphological, microscopic, and biochemical test kits (BioMerieux API, France) (CDC, 2020). Serotyping was used for further identification; and according to WHO, an antiserum set (Denka Seiken Co., Japan) is used (Ewing, 1986).
Antibacterial susceptibility test
Mueller-Hinton agar disc diffusion technology was used consisting of 12 antibiotic discs (30 µg/disc) with the following antibiotics: Gentamicin, Streptomycin, Ampicillin, Penicillin, Cefepim, Cefotaxim, Ciprofloxacin, Flumequine, Trimethophrim, Sulfametoxazole, Tetracycline and Doxycycline (Alderman and Smith, 2001).
Serological identification of E. coli by slide agglutination test
The test involved polyvalent and monovalent E. coli standard anti-serum (Li et al., 2013) that defines the enteric pathogenic type as follows: Emulsified preparation of microbial colonies was applied as two drops on a glass slide. Looped antiserum was added, aggregated, and another colony was cultivated on nutrient gradient agar and then incubated at 37°C for 24 h to test monovalent serum. A suspension of microorganisms in physiological saline was prepared and a slide agglutination test was performed to identify the antigen.
Nucleic acid extraction
DNA was extracted with the GeneJet Genomic DNA Purification Kit (ThermoFisher Scientific, USA). In summary, the microbial colonies were centrifuged for 10 min; 5000 µg of the cells pellet was resuspended in 180 µl solution for digestion (included in the kit), 20 µl K Proteinase was then added, mixed well, and incubated in a water bath at 56°C for about 30 min while shaking continuously until dissolution was complete. Further, vortex 20 µl RNase solution was added to the mixture and incubated for 10 min at 37°C. Also, 200 µl of the mixture was added to the solution, plus vortex 400 µl of 50% ethanol. The lysed cells were transferred, purified and then centrifuged for 1 min/6000 µg. Washing column was prepared by 500 µL buffer (I&II) and then re-centrifuged at maximum speed about 2 min until ethanol was completely removed. The purified DNA was stored at 20°C. Preparation of the plasmid is accomplished with the DNA plasmid GeneJet mini prep kit (Thermofisher Scientific, USA). The grown culture is placed in a 1.5 ml micro-centrifuge tube and then re-centrifuged at 12,000 xg for 2 min. The pellet is re-suspended in 250 μl of the ice-cold resuspension buffer included in the kit; thereafter, the tube is inverted approximately 56 times and mixed. Further, the tube was incubated for 5 min at 37°C. The supernatant was transferred and centrifuged at 10,000 xg/30 s, followed by rinsing of the pellet with 500 μl buffer (included in the kit) and further centrifugation at 10,000 xg for 30 s. The DNA plasmid was eluted using 50 μl of preheated double distilled H2O, incubated for 3 min at 37°C, then centrifuged again at maximum rate (14000 xg) for 30 s. For amplification of gene, a PCR reaction was used as follows: 1 μl purified gene material (genomic DNA / plasmid prep), 2.5 μl MgCl2, 5 μl buffer, 1 μl primer (as listed in Table 1), 0.25 μl enzyme mixture of Taq polymerase and 0.5 μl dNTP are combined with free nuclease water and made up to a total volume of 25 μl. Also carried out was lysis of PCR product with 0.5 µg/ml agarose gel and ethidium bromide (1%), as well as size determination of lysate using 100 bp ladder of DNA. Afterwards, the gel was run at 80 V for 50 min and documentation was done by the gel system (Biometra, Göttingen, Germany).
The DNA fragments extraction from agarose gels was as follows: DNA fragments were eluted from agarose gels by DNA Kit (Thermofisher Scientific, USA). Fragmentation is prepared by UV light and stored in a 1.5 ml tube. It was then centrifuged at 13000 xg for 2 min. Further, the column was washed using 700 μl buffer and then re-centrifuged at 1 min at 37°C. 50 µl buffer was added, eluted with a spin column filter, held at 37°C for 1 min, and then centrifuged at 13000 xg for 2 min.
RESULTS
E. coli prevalence
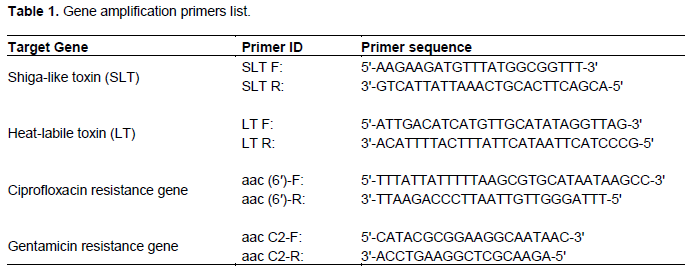
Figure 1 shows that about 100 chicken meat samples were tested on E. coli, with 40% found to be positive samples, while the remainder did not show the presence of E. coli.
Antibacterial pattern of E. coli resistance:
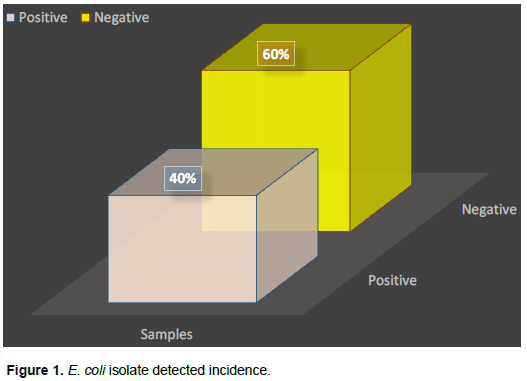
Investigation of the antimicrobial susceptibility was performed using 12 antibiotics from 6 different antibiotics families, with the test applied against twenty E. coli which were isolated from chicken samples. The obtained results were shown in Table 2. The antibiotics selected for E. coli were: the most effective antibacterial agents associated with sulfonamides; (20/20) 100% trimethoprim, 80% (16/20) sulfamethoxazole, followed by cephalosporins- about 70% (14/20) cefepime, 65% (13/20) cefotaxime; tetracycline included- 50% (10/20) tetracycline, and 40% (8/20) doxycycline. In the tested antibacterial class, the human family showed weak activity against E. coli as follows: Quinolones- (4/20) 20% ciprofloxacin, (4/20) 20% flumekin; aminoglycosides; - 3/20 (15%) gentamicin, (2/20) 10% streptomycin, and β-lactams based on the weakest member- Penicillin (10%) 2/20, and ampicillin (0%).
Serological results of E. coli isolates
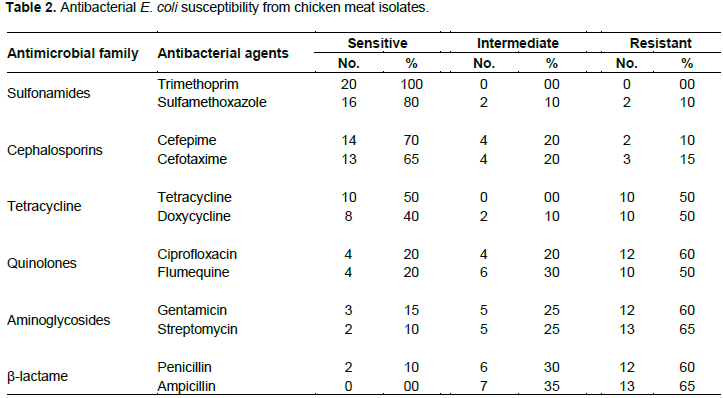
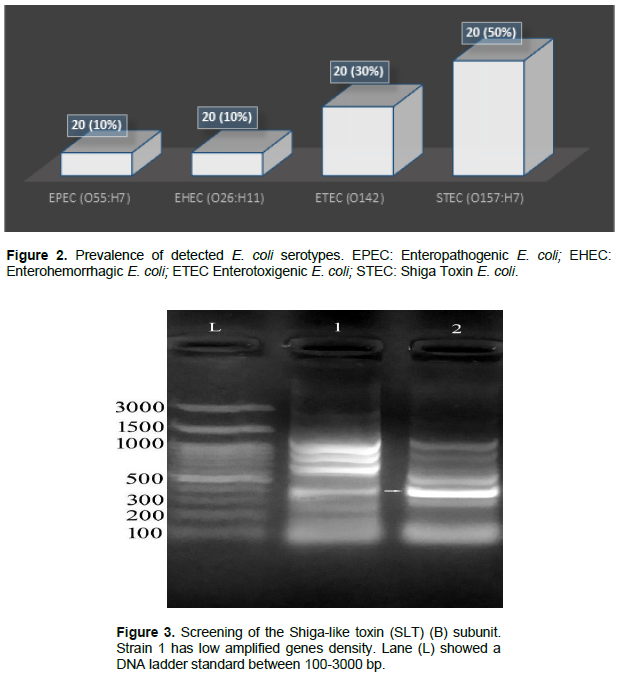
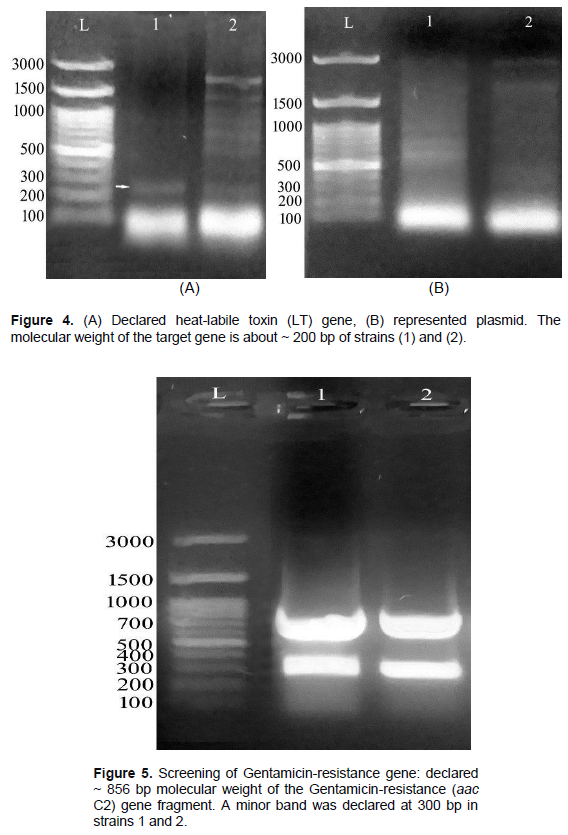
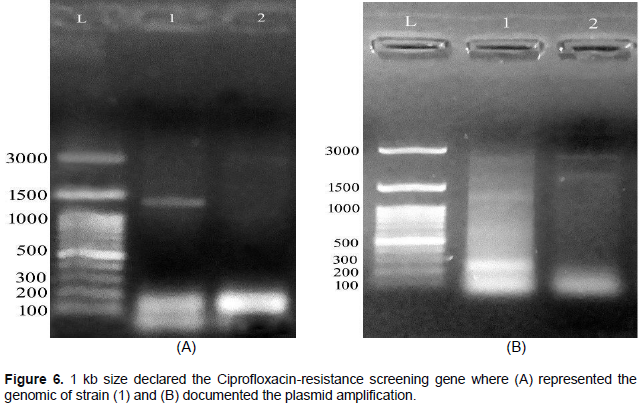
About 20 E. coli were classified serologically in Figure 2. As a result, the 10/20 (50%) isolate was STEC (O157: H7), the 6/20 (30%) isolate was ETEC (O142), and the 2/20 (10%) isolate was (O26: H11) of EHEC, 2/20 (10%) EPEC (O55: H7). The screening for SLT is described in Figure 3. Subunit B of SLT gene showed the uniform genomic DNA band with about 300 bp. of molecular weight. The results show that strain 1 had minimal amplification compared to strain 2. Fragments of approximately 200 bp were estimated in (1) and (2) strains. Figure 4A was confirmed as heat-labile toxin (LT) gene while (B) represented plasmid. The molecular weight of the target gene was about ~ 200 bp of strains (1) and (2). Gentamicin screening resistance was revealed in Figure 5 where the gentamicin gene resistance (aac C2) segment was found in the strain as a segment with a molecular weight of approximately 856 bp; however, a molecular weight of approximately 300 bp was detected. The screening for ciprofloxacin resistance is shown in Figure 6. Low Ciprofloxacin resistance genes were investigated in plasmid and genomic of tested strains. The 1 kb band was detected obviously in strain 1, but not in strain 2. In the case of the plasmid preparation, the gene target amplification was not detected.
DISCUSSION
Bacteriological investigation of 100 chicken meat samples agreed with Partridge et al. (2018) who reported 35.5% from examined Mexican chicken; Wu et al. (2018), which isolated 35.0% E. coli from chicken; Ngullie et al. (2011) who reported 31% in Indian chicken examination and Sato et al. (2010) who recorded about 20% for US chicken meat. Also, Shaltout et al. (2020) reported 13.33% E. coli from Egyptian chicken; lesser percentages of E. coli (11.1%) were reported by Tomova et al. (2018) in Nigerian chickens; Liu et al. (2016) recorded 10.60% from Croatian chicken meat; while Jakabi et. al. (2002) recorded 9% E. coli from Chinese chicken. Further, Deng et al. (2016) and Schulz et al. (2015) isolated about 5.92% of Saudi Arabian chicken meat. Lowest percentage was recorded by Collins (2000) who reported about 1.56% from Moroccan chicken. This indicates suboptimal hygiene practices at the various stages such as slaughter, handling practices, transporting, and during meat processing; leading to the presence of this microbe in processed chicken meat-and bone meal (Schulz et al., 2015). E. coli is also found in animals and human gastro-intestinal tract. Detection of this pathogen in well prepared chicken foods indicates fecal contamination, which on the other hand, indicates the possible presence of other harmful organisms such as bacterial (Salmonella, Shigella, Campylobacter) (Collins, 2000).
Antibacterial drugs are used in prevention and/or treatment, in addition to their use as chickens’ growth promoters. The benefits were achieved when the antibacterial agent were properly selected. Antibacterial susceptibility testing against different E. coli (n ??= 20) collected from chicken meat samples showed the patent antibiotics associated with sulfonamides were 100% trimethoprim (20/20), sulfamethoxazole 80% (16/20), (14/20) 70% (cephalosporins and cefepime), (13/20) 65% cefotaxime; while in the case of tetracycline usage the results showed the following: (10/20) 50% tetracycline, and (2/20) 10% streptomycin. The weakest member based on β-lactam family was as follows: (2/20) 10% penicillin and (0%) ampicillin. The obtained results agreed with CDC (2019). Nearly same results was observed by Younis et al. (2017) who reported 100% of resistance against penicillin, 95.8% against cefepime and 94.5% amoxicillin against E. coli. Ammar et al. (2015) described E. coli antibiotics resistance as caused by the presence of plasmid genes. Adeyanju and Ishola (2014) and Bie et al. (2018) stated about 90% E. coli resistant to ampicillin, tetracycline, cephalexin, trimethoprim, sulfametozazole, streptomycin, and gentamicin.
Ramadan et al. (2016) showed multidrug resistance to aminoglycosides, tetracyclines, sulfonamides and β-lactams against E. coli. Eid and Erfan (2013) and Mohamed et al. (2014) almost informed the E. coli resistance against β-lactams while Li et al. (2020) found that E. coli was highly resistant against sulfadiazine, gentamicin, amoxicillin, sulfadiazine, ampicillin, tetracycline, ceftriaxone and chloramphenicol. Zhang et al. (2012) documented that about 60% E. coli which were isolated had resistance against fluoroquinolones whereas Tang et al. (2011) found that about 35.0, 36.8, and 34.1% of ciprofloxacin, norfloxacin, and enrofloxacin were resistant.
Serological test of 20 E. coli isolates recorded about; STEC (O157: H7)10/20 (50%), ETEC (O142) 6/20 (30%), EHEC (O26: H11) 2/20 (10%) and EPEC (O55: H7) 2/20 (10%). SLT was as follows: B subunit (SLT) gene showed uniform segment in DNA genome at 300 bp molecular weight.
The results showed in strain (1) had the less amplification detection compared to heat labile toxin (LT) screening. Fragments of approximately 200 bp were recorded in both (1) and (2) strains. The resistant gentamicin gene was (aac C2) fragment documented with a molecular weight 856 bp while the small band has 300 bp molecular weight. The resistant Ciprofloxacin genes were screened on both plasmid and genomic preparations of the tested strains. Another band was detected at about 1 kb in strain (1), but not in strain (2). In the case of the plasmid preparation, target gene was not found in the isolated strain. According to Momtaz and Jamshidi (2013), O serotypes, especially O2, O1, O8, O18, O15, O35, O88, O115, O78 and O109 were the most detected serotypes. Ying et al. (2020) isolated enteropathogenic and eaeA of E. coli gene which were nearly identical to eae genes of O157: H7; O55: H7 and EHEC. Kakoullis et al. (2019) detected SLT gene was giving false negative results. On the other hand, HECO157 was detected by the 60MDa plasmid. Lagerqvist et al. (2020) detected (SLT I, II and eaeA) genes indicating the occurrence to the EHEC (O157) strain. Yang et al. (2020) documented the Stx gene detected in EHEC strains. The virulence genes include the extra-intestinal infectious genes: (afaD8, Cdt2, cdt3, traT, eisen, bmaE, iutA, iucD). Villegas et al. (2013) detected etpD gene in ETEC strain. Further, the intestinal hemorrhage was caused by EDL933 and RIMD 0509952 while fmH gene was considered as a non-virulent gene in different E. coli strains (Momtaz and Jamshidi, 2013).
CONCLUSION
The study demonstrated the presence of E. coli pathogenic genes from chicken meat samples, including various somatic capsules and antigenic genes. Managing STEC is too essential as it poses hazards to chicken consumers. E. coli presence, mainly, in our daily meals is a public health concern and food biosafety issue. It is advisable to ensure proper hygiene measures when slaughtering, handling and/or processing chicken carcasses. It is therefore recommended that unnecessary use of antibacterial drugs in living chickens and humans be avoided to forestall the emergence of new antibacterial resistance.
CONFLICT OF INTERESTS
The authors have not declared any conflict of interests.
ACKNOWLEDGEMENT
The University of Jeddah, Jeddah, Saudi Arabia is gratefully appreciated for the financial support provided (Grant No.: UJ- 20-145-DR).
REFERENCES
|
Adeyanju T, Ishola O (2014). Salmonella and Escherichia coli contamination of poultry meat from a processing plant and retail markets in Ibadan, Oyo State, Nigeria. Springerplus 3(1):139-145. |
|
|
Alderman D, Smith P (2001). Development of draft protocols of standard reference methods for antimicrobial agent susceptibility testing of bacteria associated with fish diseases. Aquaculture 196(3-4):211-243. |
|
|
Ammar M, El-Hamid I, Eid A, El Oksh S (2015). Insight into antimicrobial resistance and virulence genes of emergent multidrug resistant avian pathogenic Escherichia coli in Egypt: How closely related are they? Review of Medical Vetinary 166(9):304-314. |
|
|
Bie Y, Fang M, Li Q, Wang Y, Xu H (2018). Identification and characterization of new resistance-conferring SGI1s (Salmonella genomic island 1) in Proteus mirabilis. Frontiers of Microbiology 9:3172. |
|
|
Centers for Disease Control and Prevention (CDC) (2019). National Center for Emerging and Zoonotic Infectious Diseases (NCEZID). |
|
|
Centers for Disease Control and Prevention (CDC) (2020). National Center for Emerging and Zoonotic Infectious Diseases (NCEZID), Division of Healthcare Quality Promotion (DHQP). |
|
|
Collins D (2000). Slaughtering and processing of livestock. Journal Agricultural Mechanization and Automation 2:393-400. |
|
|
Cunrath O, Meinel M, Maturana P, Fanous J, Buyck J, Saint P, Auguste A, Helena B, Smith S, Körner J, Dehio C, Trebosc V, Kemmer C, Neher R, Egli R, Bumann A (2019). Quantitative contribution of efflux to multi-drug resistance of clinical Escherichia coli and Pseudomonas aeruginosa strains. BioMedicine 41:479-487. |
|
|
Deng Y, Wu Y, Jiang L, Tan A, Zhang R, Luo L (2016). Multi-drug resistance mediated by class 1 integrons in Aeromonas isolated from farmed freshwater animals. Frontiers of Microbiology 7:935-400. |
|
|
Eid A, Erfan M (2013). Characterization of E. coli associated with high mortality of poultry flocks. Assiut Vetinary Medical Journal 59:51-61. |
|
|
EL-Kholy M, EL-Shinawy H, Seliem H, Zeinhom A (2020). Potential risk of some pathogens in table eggs. Journal of Veterinary Medical Research 27(1):52-65. |
|
|
Ewing H (1986). The genus Shigella, in Edwards and Ewing's Identification of Enterobacteriaceae Ewing WH (ed.), Elsevier Science Publishing Co., Inc.: New York pp. 135-172. |
|
|
Jakabi M, Gelli D, Ristori C, Paula A, Sakuma H, Lopez G (2002). Presence of Salmonella Spp and Escherichia Coli O157:H7 in Raw Meat, In São Paulo City, Brazil and Evaluation of Low Temperature (Refrigeration and Freezing) Resistance of These Bacteria. Determination of human pathogen profiles in food by quality assured microbial assays Proceedings of a final Research Coordination Meeting held in Mexico City, Mexico, IAEA (internal atomic energy agency) pp. 22-26. |
|
|
Kakoullis L, Papachristodoulou E, Chra P (2019). Shiga toxin-induced haemolytic uraemic syndrome and the role of antibiotics: a global overview. Journal of Infection 79(2):75-94. |
|
|
Lagerqvist N, Lof E, Enkirch T (2020). Outbreak of gastroenteritis highlighting the diagnostic and epidemiological challenges of enteroinvasive Escherichia coli, County of Halland, Sweden, November 2017. Eurosurveillance 25(9):1900466. |
|
|
Li R, He L, Hao L, Zhou Y, Jiang H (2013). Genotypic and Phenotypic Characterization of Antimicrobial- Resistant Escherichia coli from Farm-Raised Diarrheic Sika Deer in Northeastern China. Plos One 8(9):e73342. |
|
|
Li Y, Dai X, Zeng J, Gao Y, Zhang Z, Zhang L (2020). Characterization of the global distribution and diversified plasmid reservoirs of the colistin resistance gene mcr-9. Science Reports 10:8113. |
|
|
Liu Y, Wang Y, Walsh R, Yi X, Zhang R (2016). Spencer J. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infectious Disease 16(2):161-68. |
|
|
Mohamed A, Shehata A, Rafeek E (2014). Virulence genes content and antimicrobial resistance in Escherichia coli from broiler chickens. Veterinary Medicine International Article ID 195189, 6 p. |
|
|
Momtaz H, Jamshidi A (2013). Shiga toxin-producing Escherichia coli isolated from chicken meat in Iran: Serogroups, virulence factors, and antimicrobial resistance properties. Poultry Science 92(5):1305-1313. |
|
|
Ngullie E, Walling I, Krose M, Bhatt B (2011). Traditional Animal Husbandry Practices in Tribal States of Eastern Himalaya, India: A Case Study. Indian Journal of Animal Nutrition 28:23-28. |
|
|
Partridge S, Kwong S, Firth N, Jensen S (2018). Mobile genetic elements associated with antimicrobial resistance. Clinical Microbiology Reviews 31(4):17-21. |
|
|
Ramadan H, Awad A, Ateya A (2016). Detection of phenotypes, virulence genes and phylotypes of avian pathogenic and human diarrheagenic Escherichia coli in Egypt. The Journal of Infection in Developing Countries 10(6):584-59. |
|
|
Sato H, Takahashi T, Sumitani K, Takatsu H, Urano S (2010). Glucocorticoid generates ROS to induce oxidative injury in the hippocampus, leading to impairment of cognitive function of rats. Journal of Clinical Biochemistry and Nutrition 47(3):224-232. |
|
|
Schulz S, Stephan A, Hahn S, Bortesi L, Jarczowski F, Bettmann U (2015). Broad and efficient control of major foodborne pathogenic strains of Escherichia coli by mixtures of plant-produced colicins. Proceedings of the National Academy of Sciences 112(40):E5454- E5460. |
|
|
Shaltout F, Lamda H, Edris E (2020). Bacteriological examination of cooked meat and chicken meals. Cohesive Journal of Microbiology and Infectious Disease 3(5):1-5. |
|
|
Tang X, Tan C, Zhang X, Zhao Z, Xia X (2011). Antimicrobial resistances of extra intestinal pathogenic Escherichia coli isolates from swine in China. Microbial Pathogenesis 50(5):207-212. |
|
|
Tomova A, Ivanova L, Buschmann H, Godfrey P, Cabello F (2018). Plasmid-mediated quinolone resistance (PMQR) genes and class 1 integrons in quinolone-resistant marine bacteria and clinical isolates of Escherichia coli from an aqua cultural area. Microbial Ecology 75(1):104-112. |
|
|
Villegas N, Baronetti J, Albesa I, Polifroni R, Parma A, Etch everría A, Becerra M, Padola N, Paraje M (2013). Relevance of biofilms in the pathogenesis of Shiga-toxin-producing Escherichia coli infection. Scientific World Journal Article ID 607258, 7 p. |
|
|
Wu S, Huang J, Wu Q, Zhang J, Zhang F, Yang X, Wu H, Zeng H, Chen M, Ding Y, Wang J, Lei T, Zhang S, Xue L (2018). Staphylococcus aureus Isolated from Retail Meat and Meat Products in China: Incidence, Antibiotic Resistance and Genetic Diversity. Frontiers in Microbiology 9:2767. |
|
|
Yang X, Sun H, Fan R. (2020). Genetic diversity of the intimin gene (eae) in non-O157 Shiga toxin-producing Escherichia coli strains in China. Scientific Reports10(1):3275-3283. |
|
|
Ying H, Xiangning B, Ji Z, Cecilia J, Milan C, Sverker H, Anne F, Xi Y, Yanwen X, Chengsong W, Andreas M (2020). Molecular characteristics of eae-positive clinical Shiga toxin-producing Escherichia coli in Sweden. Emerging Microbes and Infections 9(1):2562-2571. |
|
|
Younis G, Awad A, Mohamed N (2017). Phenotypic and genotypic characterization of antimicrobial susceptibility of avian pathogenic Escherichia coli isolated from broiler chickens. Veterinary World 10(10):1167-1172. |
|
|
Zhang T, Wang C, Zhong X (2012). Survey on sulfonamide antibiotic- resistant genotype and phenotype of avian Escherichia coli in North China. Poultry Science 91(4):884-887. |
|
Copyright © 2024 Author(s) retain the copyright of this article.
This article is published under the terms of the Creative Commons Attribution License 4.0