ABSTRACT
Girardinia diversifolia (Link) Friis is a perennial herb commonly known as Himalayan giant nettle which belongs to the family Urticaceae. The plant has cultural, medicinal and economic importance. The plant contains high concentration of polysaccharides, polyphenols and secondary metabolites which obstructs the process of isolation of the Deoxyribonucleic Acid (DNA) and inhibits downstream Polymerase Chain Reaction (PCR) amplifications. This study protocol developed a DNA extraction protocol from leaf-tissue based on the Cetyltrimethylammonium bromide, and optimized the PCR protocol for Inter Simple Sequence Repeat (ISSR) analysis. Genomic DNA extraction process was conducted using modified Doyle and Doyle method to obtain good quality DNA. The method yielded 445 ng/µL of DNA, where the purity ranged from 1.8-2.0 indicating minimum contamination of metabolites. The optimum condition for ISSR analysis was established using 4 mM MgCl2 , 0.6 mM dNTPs, 2.0 U Taq polymerase, 50 ng template DNA, and 0.7 µM primer. PCR program was optimized in the sequence of denaturation at 94°C for 3 min, subsequently followed by 45 cycles at 94°C for 30 s, annealing temperature at 45°C for 30 s, extension at 72°C for 2 min, and final extension at 72°C for 10 min. The modified technique was found to be ideal for isolation of genomic DNA and optimization of PCR process for ISSR analysis of G. diversifolia. The results of the research are beneficial for future molecular characterization and genetic diversity analysis of allied taxa.
Key words: Girardinia diversifolia, Himalayan nettle, DNA isolation.
Girardinia diversifolia (Link) Friis is commonly known as Himalayan giant nettle and locally known as ‘Allo’ in Nepal. The plant belongs to family Urticaceae which contains approximately 54 genera and has more than 2000 species with high concentration of genera and species in tropical Asia (Wu et al., 2013). This plant is widely distributed in the subtropical and temperate Himalayas (Polunin and Stainton, 1984) and its habitat is found between the altitudes of 1,200 to 3,000 metres above sea level (Friis, 1981; Shrestha, 1997; Singh and Shrestha, 1988). Molecular genetic markers reflect the variation at the level of DNA (Anne, 2006). Inter Simple Sequence Repeat (ISSR)-PCR is a technique, which utilizes microsatellite sequence in polymerase chain reaction to generate multilocus markers (Reddy et al., 2002). This technique with great reproducibility has been widely used in classification and systematic comparison of species (Amom et al., 2018; Nilkanta et al., 2017), evolutionary relationship of species and identification of genetic varieties (Mohamad et al., 2017).
Himalayan giant nettle has great cultural, economic and medicinal significance among the indigenous people and local communities (IPLCs) of Nepal (Barakoti and Shrestha, 2008; Subedee et al., 2020). Fiber obtained from the stem of this plant is used to make clothes, fishing nets, bags, coats and many other textile products. The plant is used in traditional medicine for treating gastritis, joint pain, headache, and skin allergies (Barakoti and Shrestha, 2008; Subedee et al., 2020). Market demand of the products made from this plant is increasing year by year, and created a risk of over exploitation from its natural habitat. Generic phylogeny and character evaluation in Urticaceae including G. diversifolia from China has been studied based on analysis of nuclear ribosomal internal transcribed spacer (nrITS) and two plastid DNA regions rbcL exon and trnL-F spacer (Wu et al., 2013). However genetic diversity of G. diversifolia by using ISSR has not been conducted yet. Thus, there is a need to study molecular characteristics of this plant by using ISSR.
G. diversifolia contains many bioactive compounds such as β-sitosterol, 7- hydroxysitosterol and 3-hydroxystigmast-5-en-7-one (Njogu et al., 2011)trans syringin, linoleic and linolenic acid (Shrestha et al., 2020). Genetic analysis of plants provides broader knowledge on their diversity and basis to study important metabolites produced by them (Fernie and Klee, 2011). Evaluation of genetic diversity in plants requires high quality and quantity of DNA (Sá et al., 2011)for which G. diversifolia presented a great challenge. Secondary metabolites present in plants can interfere in genomic DNA extraction, purification and downstream applications (Friar, 2005). Urticaceae family members produce large amounts of exudate, which obstruct DNA extraction (Sarrazola and Alzate, 2019). Various methods of DNA extraction has been carried out on many species of Urticaceae family (Bharmauria et al., 2009; Wu et al., 2013)which has indicated that further modifications are essential to obtain good quality genomic DNA for PCR based analysis (Aboul-Maaty and Oraby, 2019). Despite high value, very few information is available on extraction of its genomic DNA. Isolation of DNA from leaf tissue was found to contain high degree of phytochemical while using standard protocol by Doyle and Doyle (1987). These phytochemicals inhibited PCR reaction. Hence, modification in the extraction protocol was a requisite to obtain good quality DNA. The objective of this study is to develop DNA extraction protocol and optimization of PCR protocol for ISSR-PCR analysis of G. diversifolia.
Plant material
Seeds of G. diversifolia were collected from Naugad rural municipality of Darchula District, Far-western region of Nepal (29°47’34.9”N, 80°36’23.5”E). The seeds were thoroughly rinsed with distilled water, allowed to germinate on top of moist absorbent paper in plastic petri dishes using top-of-paper method (Rao et al., 2006) and transplanted to small pot after 12 days. The germinated plants were maintained at Truffle Research Centre, Coronation Garden of Tribhuvan University, Kirtipur, Kathmandu, Nepal (27°40’50”N, 85°17’26.5”E). Young fresh juvenile leaves of the plants were collected prior to extraction of DNA.
Reagents used in isolation of genomic DNA
Cetyltrimethylammonium bromide (CTAB) was modified which consists of extraction buffer 2% (w/v) CTAB (Sigma, Sintra, Portugal), 1 M Tris Hcl (pH 8.0), 0.5 M EDTA (pH 8.0), 5 M NaCl, with 5% PVP (w/v) Polyvinlypyrrolidone (Sigma, Sintra, Portugal), 5% β-mercaptoethanol, Ammonium acetate 7.5 M, (25:24:1) Phenol:Chloroform:Isoamyl alcohol (Sigma-aldrich). Ethanol 70% and 100% were used for isolation of DNA.
Isolation of genomic DNA
Freshly harvested young leaves (100 mg) were ground in liquid nitrogen into fine powder with the help of mortar and pestle. The study followed CTAB, DNA isolation protocol of Doyle and Doyle (1987), and modified the DNA extraction method. The modification of Doyle and Doyle protocol to extract genomic DNA was followed as described below.
Prior to DNA extraction, 5% β-mercaptoethanol and 5% PVP (Polyvinylpyrrolidone) was added in CTAB buffer. Freshly harvested young leaf samples (100 mg) were ground in liquid nitrogen in a pre-chilled mortar and pestle. CTAB buffer (500 µL) was added quickly and transferred into sterile 1.5 mL centrifuge tubes. The tubes were incubated at 55°C for 1 h with occasional shaking for every 10 min. Phenol:chloroform:isoamyl alcohol (25:24:1) was mixed well (500 µL) to form an emulsion by shaking tubes. Centrifugation was carried out at 14000 Revolution Per Minute (RPM). Phenol:chloroform:isoamyl alcohol step was repeated twice. The supernatant was carefully decanted and transferred to new tubes, pre-chilled 0.08 volumes of 7.5 M Ammonium acetate and 0.54 volumes of cold isopropanol was added and mixed well.
The samples were kept in -20°C for 1 h and centrifuged at 14000 RPM for 3 min. The pellet was washed twice with 70% and once in 100% ethanol. The supernatant was decanted and DNA pellet was air-dried at room temperature until the white pellet turned transparent. The DNA pellet was resuspended in 100 µL of TE buffer.
Quantification of extracted DNA and testing for purity
The yield of extracted DNA was measured in a nanospectrometer (Bio-spec nano) at 260 nm. The purity of DNA was measured by estimating the ratio of absorbance at 260 to 280 nm. The DNA purity was determined by running the sample in 0.8% agarose gel.
The size of each fragments was estimated using 100 bp plus DNA ladder.
Optimization of Polymerase Chain Reaction (PCR)
The optimization of ISSR-PCR was carried out with the extracted genomic DNA from G. diversifolia. Five major factors; Taq DNA polymerase (Vivantis, Malaysia), dNTPs (Vivantis, Malaysia), primers (University of British Columbia, Canada), MgCl2 (Vivantis, Malaysia), template DNA, and their concentrations were considered highly as shown in Table 4. The ISSR primer obtained from University of British Columbia (UBC) were used for optimization of PCR condition.
The reaction was carried out in the DNA thermocycler (Biorad T100). Total volume of 15 µL PCR reaction mixture was used which contained 1X buffer, (0.1-0.6) mM dNTPs, (1.5-4) mM Mgcl2, (0.1-0.9) µM primer, (0.5-3.0) unit Taq polymerase and (25-100) ng of DNA template. The thermocycler was programmed and optimized by testing various conditions: 3 min at 93°C, followed by 45 cycles for 30 s at 93°C, 45 s at different annealing temperature (45-52)°C, extension at 72°C for 2 min, final extension at 72°C for 10 min and finally holding temperature at 4°C. After PCR reaction, electrophoresis of The PCR products were carried out in 1.8% agarose gel containing 10 mg/mL of ethidium bromide, 1X TAE buffer at 80 V for 1 h. DNA ladder of 100 base pair was used for determining the molecular weight. The DNA bands were observed under ultraviolet light using Gel documentation system.
Isolation and purity detection of DNA
The mean concentration and purity of DNA samples extracted from the leaves of G. diversifolia are presented in Table 2.
Yield of DNA from Doyle and Doyle method leaf tissue was found to have high degree of phytochemical and thus needed modification of the method. The modified method yielded 445 ng/µL of DNA, the purity ranged between 1.8 -2.0 indicating minimum contamination of metabolites.
High concentration of β-mercaptoethanol and PVP in extraction buffer played a major role in neutralization of polyphenols, tannins and oxidation of secondary metabolites (Porebski et al., 1997). The modified protocol showed high efficiency to extract good quality DNA by varying the concentration of NaCl, β-mercaptoethanol and PVP. The modified conditions are listed in Table 3.
Optimization of PCR-ISSR and screening primers
For optimization of ISSR-PCR, various concentrations were considered including template DNA, primers, Taq polymerase, dNTPs, MgCl2, annealing temperature. The optimized conditions for ISSR-PCR protocol are given in Table 4.
The optimized condition for ISSR-PCR reaction for G. diversifolia was found to be 50 ng of genomic DNA, 4 mM Magnesium chloride, 0.6 mM dNTPs, 0.7 µM primer concentration, 2 U Taq polymerase in total 15 µL PCR reaction volume. The reproducible clear bands in agarose gel electrophoresis is shown in Figure 1.
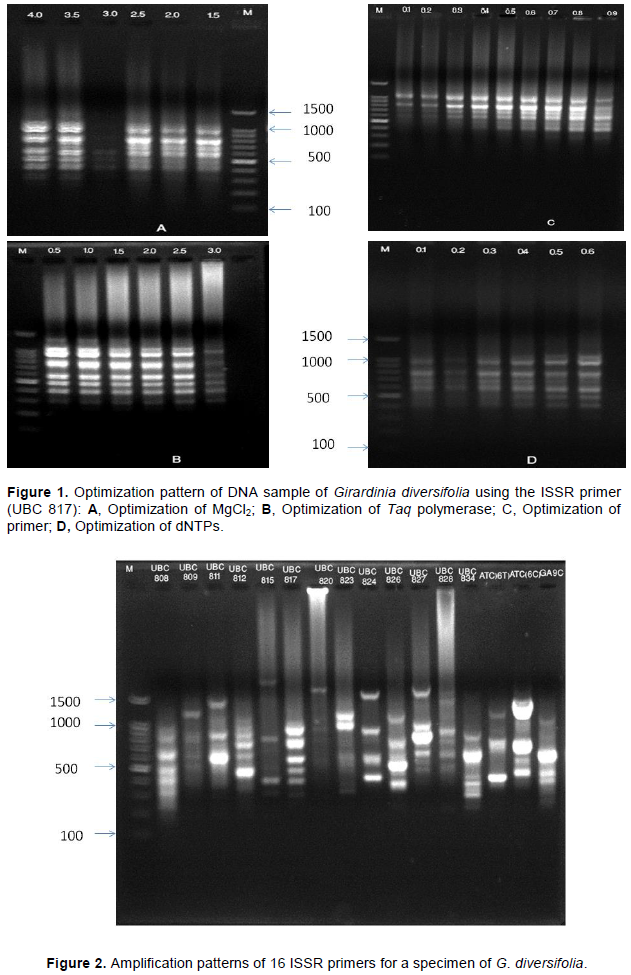
DNA extracted from G. diversifolia using the optimized PCR parameters are shown in Table 4. Among 16 screened ISSR primers (Table 1), seven ISSR primers (UBC811, UBC812, UBC817, UBC824, UBC826, UBC827, UBC 834) showed the clear, reproducible bands and the experiment was repeated twice (Figure 2). Total volume of 15 µL PCR reaction mixture which contained 1× buffer, 0.6 mM dNTPs, 4 mM Mgcl2, 0.7 µM primer, 2 unit Taq polymerase and 50 ng of DNA template were optimized (Figure 1 and Table 4). The thermocycler was optimized by testing conditions: 3 min at 93°C, followed by 45 cycles for 30 second at 93°C, 45 second at different annealing temperature 45°C, extension at 72°C for 2 min, final extension at 72°C for 10 min and finally holding temperature at 4°C. After PCR reaction, electrophoresis of the PCR products were carried out in 1.8% agarose gel containing 10 mg/mL of ethidium bromide, 1× TAE buffer at 80 V for 1 h. DNA ladder of 100 basepair was used for determining the molecular weight. The optimal ISSR-PCR reaction conditions were used for the study of genetic diversity of G. diversifolia separately.
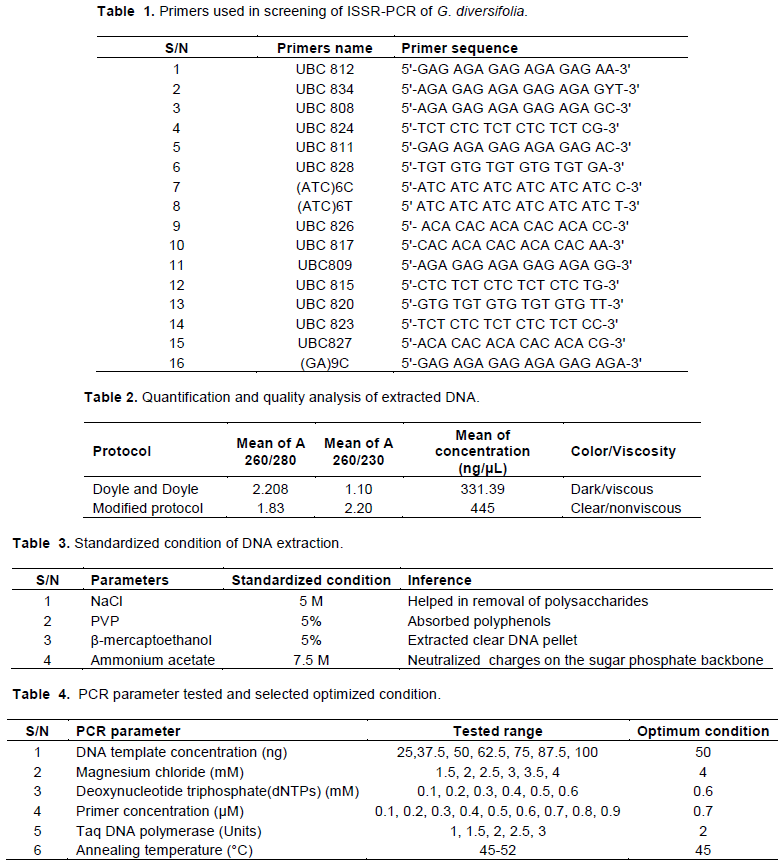
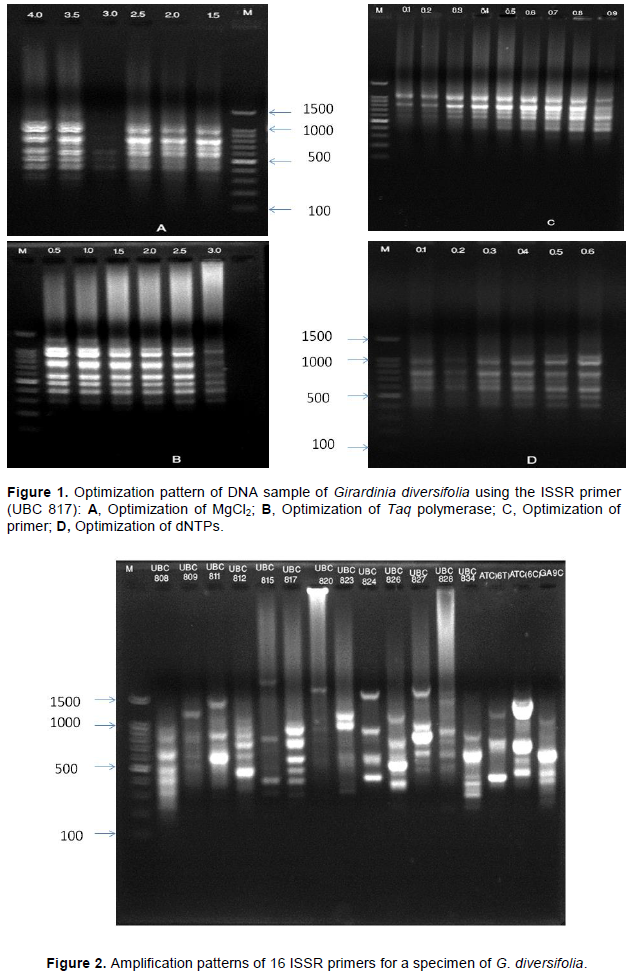
Genomic DNA isolation from G. diversifolia using Doyle and Doyle (1987) standard protocol was conducted. Many obstacles were encountered from the very first step of genomic DNA extraction, including cell lysis, DNA elution and subsequent PCR reactions. The biochemical composition in plant tissues of different species is expected to vary considerably and may not yield optimal DNA from one isolation protocol (Khanuja et al., 1999; Kumari et al., 2020). Due to the presence of PCR inhibitors, protocol of Doyle and Doyle (1987) did not show good quality DNA. It resulted in the absence of DNA band in electrophoresis technique. Thus, procedure described by Doyle (1987) was modified by altering the parameters: increased concentration of NaCl from 2 to 5M, increased concentration of β-mercaptoethanol, increased concentration PVP, addition of phenol to the deproteinization process.
The recommended values for the A260/A280 ratio ranged from 1.8-2.0 and absorption ratio at A260/230 is 2.0-2.22 for impurity free DNA (Arruda et al., 2017). Higher value of absorbance from 2.0 indicated the contamination of phenol in extracted DNA while lower value indicated the presence of proteins. The optimized protocol presented in the study showed a mean DNA concentration of 400 ng/µl extracted from the leaf of G. diversifolia. The method resulted in the mean value of 1.83 which confirmed the extraction of pure DNA at A260/A280 and A260/A230 ratios.
High concentration of NaCl (5 M) effectively removed polysaccharides during DNA extraction of G. diversifolia (Table 3). Studies in other plants species like Mangifera indica, Capsicum sp. Eclipta alba, Aegle marmelos, Grewia asiatica, supported that this modification allowed an efficient elimination of polysaccharides (Devi et al., 2018; Kit and Chandran, 2010; Kumar et al., 2018; Mujeeb et al., 2017; Shukla et al., 2018). High concentration of NaCl helped in the elimination of polyphenols from the leaf of G. asiatica (Shukla et al., 2018).
Use of high concentration of PVP improved the quality of DNA by removing secondary metabolites during genomic DNA extraction process (John, 1992; Osena et al., 2017). The modified protocol used 5% PVP in 2% CTAB buffer which was effective in the elimination of polyphenols that resulted in the extraction of clear DNA pellets. Some studies also suggested the use of 5% PVP in CTAB buffer Vigna sp. (Choudhary et al., 2008)and Mimosa tenuiflora (Arruda et al., 2017)to obtain good quality DNA pellet.
β-mercaptoethanol enhances denaturation of protein (Tiwari et al., 2012). In the modified protocol, the concentration of β-mercaptoethanol was increased to 5%. The high concentration of β-mercaptoethanol is important for the reduction of polyphenols during extraction of genomic DNA of plants containing high content of secondary metabolites (Arruda et al., 2017). The modified protocol contains 5% β-mercaptoethanol instead of 2% as used by Doyle and Doyle (1987). The modified protocol helped to reduce brown coloured DNA pellet. Other study carried out in Litchi chinensis (Arruda et al., 2017; Puchooa, 2004)also reported that increase in the concentration of β-mercaptoethanol helped to extract clear DNA pellet. Polyphenols often damage extracted genomic DNA and make some enzymes inaccessible (Anerao et al., 2016). The use of phenol: choloroform: isoamyl alcohol effectively removed polyphenols and yielded pure genomic DNA.
For the identification of polymorphic characteristics and genetic diversity of plant species, ISSR acts as a powerful method since it depends on the quality and quantity of extracted DNA. Selection of primers is an important factor because the same primer may exhibit different amplification results in different species.
The Doyle and Doyle (1987) protocol was successfully modified to isolate genomic DNA by increasing the concentration of PVP, β-mercaptoethanol, NaCl and addition of Phenol:chloroform:isoamyl alcohol (25:24:1) in the extraction buffer. These changes made it possible to obtain high quality genomic DNA from G. diversifolia. The extracted DNA was used to optimize PCR based ISSR protocol which gave clear and amenable DNA bands. The obtained results confirm that the modified protocol is suitable with G. diversifolia and other plant species containing high concentration of secondary metabolites. The research is beneficial for future molecular characterization, genetic diversity analysis of allied taxa and genetic improvement works.
This study was supported by Kailash Sacred Landscape Conservation and Development Initiative (KSLCDI) Nepal, a collaborative programme between Ministry of Forests and Environment, Government of Nepal; Research Centre for Applied Science and Technology, Tribhuvan University; and International Centre for Integrated Mountain Development (ICIMOD); the German Federal Ministry for Economic Cooperation and Development (BMZ) through the Deutsche Gesellschaft für Internationale Zusammenarbeit (GIZ) GmbH and the United Kingdom’s Department for International Development (DFID) – UK Aid, as well as by the core funds of ICIMOD contributed by the governments of Afghanistan, Australia, Austria, Bangladesh, Bhutan, China, India, Myanmar, Nepal, Norway, Pakistan, Switzerland, and the United Kingdom. The views and interpretation in this publication are those of the authors and should not be ascribed to MoFE, RECAST, ICIMOD or their donors.
The authors have not declared any conflict of interests.
REFERENCES
|
Aboul-Maaty NF, Oraby HAS (2019). Extraction of high-quality genomic DNA from different plant orders applying a modified CTAB-based method. Bulletin of the National Research Centre 43(1):25.
Crossref
|
|
|
|
Amom T, Tikendra L, Rahaman, Nongdam P (2018). Evaluation of genetic relationship between 15 bamboo species of North-East India based on ISSR marker analysis. Molecular Biology Research Communications 7(1):7-15.
|
|
|
|
|
Anerao J, Jha V, Desai N (2016). Optimization of DNA Extraction Methods from Garcinia species for ISSR-PCR, RAPD-PCR and DNA Barcoding. Asian Journal of Biotechnology 9(1):35-42.
Crossref
|
|
|
|
|
Anne C (2006). Choosing the right molecular genetic markers for studying biodiversity: From molecular evolution to practical aspects. Genetica 127:101-120.
Crossref
|
|
|
|
|
Arruda SR, Pereira DG, Silva-Castro MM, Brito MG, Waldschmidt AM (2017). An optimized protocol for DNA extraction in plants with a high content of secondary metabolites, based on leaves of Mimosa tenuiflora (Willd.) Poir. (Leguminosae). Genetics and Molecular Research 16(3):1-9.
Crossref
|
|
|
|
|
Barakoti TP, Shrestha KP (2008). Commercial utilization of Allo (Girardinia diversifolia) by the Rais of Sankhuwasabha for income generation. Banko Janakari 18(1):18-24.
Crossref
|
|
|
|
|
Bharmauria V, Narang N, Verma V, Sharma S (2009). Genetic variation and polymorphism in the Himalayan nettle plant Urtica dioica based on RAPD marker. Journal of Medicinal Plants Research 3(3):166-170.
|
|
|
|
|
Choudhary K, Mathur N, Choudhary OP, Pillai U (2008). Protocol for Isolation of Genomic Dna from Dry and Fresh Leaves of Vigna Species Suitable for Rapd and Restriction Digestion. Advances in Biological Research 2(5-6):83-89.
|
|
|
|
|
Devi AA, Brajendra N, Dinachandra M (2018). Genetic Diversity Analysis in Chilli (Capsicum annuum L.) Found in Manipur Using RAPD Markers. International Journal of Current Microbiology and Applied Sciences 7(10):257-262.
Crossref
|
|
|
|
|
Doyle JJ, Doyle JL (1987). A Rapid DNA Isolation Procedure for Small Quantities of Fresh Leaf Tissue. Phytochemical Bulletin 19:11-15.
|
|
|
|
|
Fernie AR, Klee HJ (2011). The Use of Natural Genetic Diversity in the Understanding of Metabolic Organization and Regulation. Frontiers in Plant Science 2:59.
Crossref
|
|
|
|
|
Friar EA (2005). Isolation of DNA from Plants with Large Amounts of Secondary Metabolites. In Methods in Enzymology 395:1-12.
Crossref
|
|
|
|
|
Friis I (1981). A Synopsis of Girardinia (Urticaceae). Kew Bulletin 36(1):143-157.
Crossref
|
|
|
|
|
John ME (1992). An efficient method for isolation of RNA and DNA from plants containing polyphenolics. Nucleic Acids Research 20(9):2381-2381.
Crossref
|
|
|
|
|
Khanuja SP, Shasany AK, Darokar MP, Kumar S (1999). Rapid isolation of DNA from dry and fresh samples of plants producing large amounts of secondary metabolites and essential oils. Plant Molecular biology Reporter 17(1):74.
Crossref
|
|
|
|
|
Kit YS, Chandran S (2010). A simple, rapid and efficient method of isolating DNA from Chokanan mango ( Mangifera indica L.). African Journal of Biotechnology 9(36):5805-5808.
|
|
|
|
|
Kumar A, Singh I, Badari NS (2018). Extraction of Genomic DNA from Dried mature leaves of Eclipta alba (L.) suitable for ISSR-PCR and other DNA fingerprinting and Barcoding techniques. International Journal of Molecular Biotechnology 4(2):23-30.
|
|
|
|
|
Kumari M, Jadhav AS, Chahande RV (2020). Isolation of genomic DNA from groundnut plant by modified, rapid and efficient protocol. Journal of Pharmacognosy and Phytochemistry 9(1):2268-2271.
|
|
|
|
|
Mohamad A, Alhasnawi AN, Kadhimi AA, Isahak A, Wan Yusoff WM, Che Radziah CMZ (2017). DNA Isolation and Optimization of ISSR-PCR Reaction System in Oryza sativa L. International Journal on Advanced Science, Engineering and Information Technology 7(6):2264.
Crossref
|
|
|
|
|
Mujeeb F, Bajpai P, Pathak N, Verma SR (2017). Genetic Diversity Analysis of Medicinally Important Horticultural Crop Aegle marmelos by ISSR Markers. In L. Domingues (Ed.), PCR. Springer New York. pp. 195-211.
Crossref
|
|
|
|
|
Nilkanta H, Amom T, Tikendra L, Rahaman H, Nongdam P (2017). ISSR Marker Based Population Genetic Study of Melocanna baccifera (Roxb.) Kurz: A Commercially Important Bamboo of Manipur, North-East India. Scientifica 2017:1-9.
Crossref
|
|
|
|
|
Njogu PM, Thoithi GN, Mwangi JW, Kamau FN, Kibwage IO, Kariuki ST, Mwalukumbi JM (2011). Phytochemical and Antimicrobial Investigation of Girardinia diversifolia (Link)Friis (Urticaceae). East and Central African Journal of Pharmaceutical Sciences 14(3):89-94.
|
|
|
|
|
Osena G, Nyaboga E, Amugune N (2017). Rapid and Efficient Isolation of High Quality DNA from Cassava (Manihot esculenta Crantz) Suitable for PCR Based Downstream Applications. Annual Research and Review in Biology 12:1-10.
Crossref
|
|
|
|
|
Polunin O, Stainton A (1984). Flowers of the Himalaya. Oxford University Press.
|
|
|
|
|
Porebski S, Bailey LG, Baum BR (1997). Modification of a CTAB DNA extraction protocol for plants containing high polysaccharide and polyphenol components. Plant Molecular Biology Reporter 15(1):8-15.
Crossref
|
|
|
|
|
Puchooa D (2004). A simple, rapid and efficient method for the extraction of genomic DNA from lychee (Litchi chinensis Sonn.). African Journal of Biotechnology 3(4):253-255.
Crossref
|
|
|
|
|
Reddy MP, Sarla N, Siddiq EA (2002). Inter simple sequence repeat (ISSR) polymorphism and its application in plant breeding. Euphytica 128(1):9-17.
Crossref
|
|
|
|
|
Sá O, Pereira JA, Baptista P (2011). Optimization of DNA Extraction for RAPD and ISSR Analysis of Arbutus unedo L. Leaves. International Journal of Molecular Sciences 12:4156-4164.
Crossref
|
|
|
|
|
Sarrazola JH, Alzate FA (2019). Obtaining DNA from Urticaceae: Overcoming the challenges associated with chemical compounds and herbarium specimens. International Journal of Molecular Biology 4(5):158-165.
Crossref
|
|
|
|
|
Shrestha SS, Sut S, Ferrarese I, Barbon Di Marco S, Zengin G, De Franco M, Pant DR, Mahomoodally MF, Ferri N, Biancorosso N, Maggi F, Dall Acqua S, Rajbhandary S (2020). Himalayan Nettle Girardinia diversifolia as a Candidate Ingredient for Pharmaceutical and Nutraceutical Applications-Phytochemical Analysis and In Vitro Bioassays. Molecules 25:1563.
Crossref
|
|
|
|
|
Shrestha R (1997). Cytological Studies in Girardinia Diversifolia (Link) Friis. Pakistan Journal of Botany 29(2):263-269.
|
|
|
|
|
Shukla R, Sharma DC, Pathak N, Bajpai P (2018). Genomic DNA Isolation from High Polyphenolic Content Grewia asiatica L. Leaf Without Using Liquid Nitrogen. Iranian Journal of Science and Technology, Transactions A: Science 42:347-351.
Crossref
|
|
|
|
|
Singh SC, Shrestha R (1988). Girardinia diversifolia (Urticaceae), a non-conventional fiber resource in Nepal. Economic Botany 42:445-447.
|
|
|
|
|
Subedee BR, Chaudhary RP, Uprety Y, Dorji T (2020). Socio-ecological perspectives of Himalayan Giant Nettle ( Girardinia diversifolia (Link) Friis) in Nepal. Journal of Natural Fibers 17(1):9-17.
Crossref
|
|
|
|
|
Tiwari KL, Jadhav SK, Gupta S (2012). Modified CTAB Technique for Isolation of DNA from some Medicinal Plants. Research Journal of Medicinal Plant 6(1):65-73.
Crossref
|
|
|
|
|
Wu ZY, Monro AK, Milne RI, Wang H, Yi TS, Liu J, Li DZ (2013). Molecular phylogeny of the nettle family (Urticaceae) inferred from multiple loci of three genomes and extensive generic sampling. Molecular Phylogenetics and Evolution 69(3):814-827.
Crossref
|
|